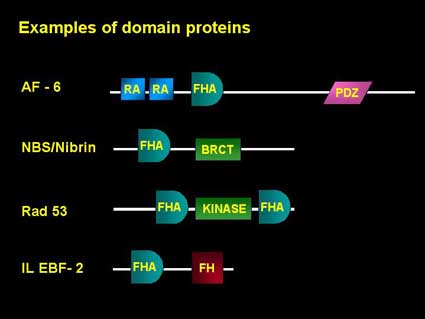
 Structure:
Structure:
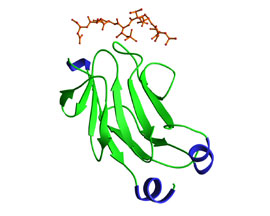
The FHA domain forms an 11-stranded b-sandwich that has a short a-helix inserted between b strands 2 and 3 and an alpha-helical region at the extreme C-terminus. The b-strand topology is essentially identical to that of the MH2 domain of Smad proteins, although the a-helical insertions involved in MH2 receptor binding and homo-oligomerization differ extensively. The N- and C-termini are in close proximity to one another since the first and last b strands in the sandwich lie adjacent to one another. The peptide binding site is located on the opposite side of the FHA domain, an arrangement that is consistent with the modular nature of an independent folding unit. The phosphothreonine peptide binds at a site created by the loop regions between b 3/4, b4/5, and b6/7, while regions not involved in creating the structural basis for these loops contain variable insertions and share little homology with one another. The figure presents the first FHA domain of Rad53 in complex with a phosphothreonine containing peptide. In this structure, the primary contact on the peptide occurs at the phosphothreonine and the +4 aspartic acid residue.
Reference: Durocher et al. 2000. Mol. Cell 6 (5):1162-1182.