
Back to "Introduction to biological significance of bioinformatics topics"
Back to "Topics"
The molecular basis of disease, mimicry and functional design - using disease as example. Disease can in the following be replaced by mimicry and functional design without much adjustments.
Introduction:
- living organisms as molecular circuitry:
intended modes of operation = healthy state
aberrant modes of operation = disease state
- Diagnosis:
identify the molecular basis of disease
- Therapy:
guide biochemical circuitry back to healthy state
Molecular circuitry:
- biochemical processes, that form and recycle molecules in a coordinated and balanced fashion
- simple view of one area within the complex network:

Figure: A metabolic network. Overview of metabolism as a molecular circuit. Taken from www.genome.ad.jp/kegg/kegg.html on September 23, 2002.
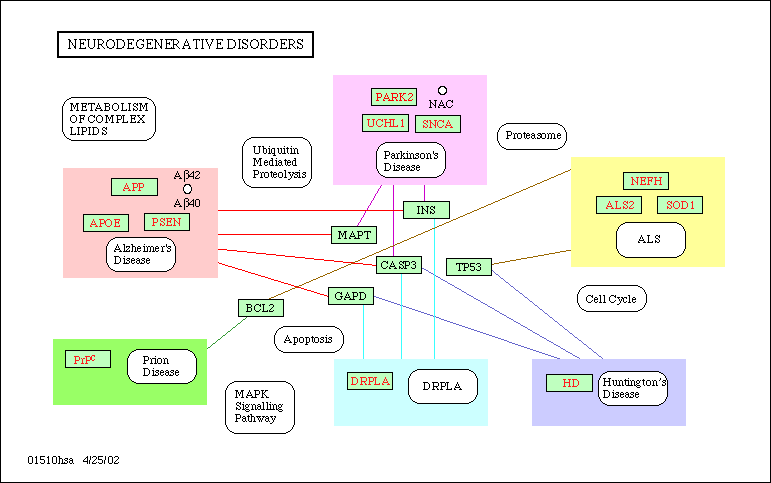
Figure: A regulatory network. Overview of neurodegenerative disorders in humans. Taken from www.genome.ad.jp/kegg/kegg.html on September 23, 2002.
Explanation of Graph representation in metabolic network:
dot (node) is a small molecule that is metabolized
line (edge) is a chemical reaction. This reaction can be catalyzed by an enzyme, and may require a co-factor and/or other reaction partners. The enzyme may be inhibited or activated by regulatory mechanisms. Enzymes are proteins. Example:

Figure: In a metabolic pathway map such as shown above, each node and line correspond to a small molecule and a chemical reaction, respectively. Here they are shown in blue.
View of disease in this molecular circuitry description:
The molecular basis of disease lies in modification of one or more components of the metabolic or regulatory networks. An imbalance in just one of the components of a circuit, can throw the entire circuit off balance. Examples:
- reactions do not happen at their normal rate (sickle cell anemia, diabetes)
- resources are not present in sufficient amounts (vitamin deficiencies)
- waste products accumulate in the body (misfolding diseases, such as Alzheimer's)
Molecular approach to curing diseases:
Design drugs that regulate proteins.
Examples:
- inhibit an enzyme by providing a drug molecule that effectively competes with the natural substrate of the enzyme (inhibitor). Requirements: drug must bind more strongly to enzyme than substrate.
- bind to receptor molecules who are located in the cell membrane and fulfill regulatory tasks
- bind to ion channels and transporter systems that monitor the flux of molecules in and out of the cell
Tasks within this approach:
1. To study molecular details of a function or condition
2. From 1, to find a protein target for a disease
3. To design the drug that binds to the protein target
Generalization to other aspects but disease:
1. same as disease
2-3. will be different, e.g. instead of designing a drug, it could be adding a subunit, or modifying an existing protein etc.
1. Studying molecular details of function or condition experimentally and role of bioinformatics
- traditionally not systematic, new drugs based on modification of old drugs
- all drugs on the marked world-wide target 500 proteins (Drews, J. (1998) Die verspielte Zukunft. Basel, Switzerland, Birkhaueser Verlag.)
=> search for protein targets is the bottleneck in pharmaceutical research
- today: systematic approaches:
Genomics
A. DNA chips B. Whole-genome comparison
- compare normal versus disease state - compare virulent versus non-virulent strains
Proteomics
whole-cell protein gels, followed by mass spectrometry
Pros and Cons Genomics versus Proteomics
Genomics is experimentally easy, Proteomics is experimentally difficult and some proteins are more difficult than others.
Proteomics provides more information. For example, Genomics does not, but Proteomics does give information on posttranslational modifications.
Genomics doesn't give direct information on protein levels, since they can be very different from expression levels, while proteomics does
Both, genomics and proteomics do not provide higher level information on the structure, dynamics, function of the proteins (e.g. location, place and role in metabolic or regulatory networks etc.).
Role of Bioinformatics:
help with information that is not provided by genomics and proteomics approaches: higher level information
=> piece together all available information
1. to get detailed picture of a molecular process (or disease)
2. from 1, to identify new protein targets
3. from 2, to develop drugs ("cheminformatics")
a. based on chemical similarity of known drugs
b. rational (structure-based) drug design interactively on computer screen
c. molecular docking (automatic, systematic computer-based prediction of structure and binding affinity of complex)
d. high-throughput screening and combinatorial chemistry
Components of Bioinformatics that contribute to understanding molecular processes (and thereby finding protein targets, etc): 7 hierarchical layers
Layer 1. Sequencing support
challenges: base calling, physical mapping, fragment assembly
outcome: raw genome sequence
Layer 2. DNA sequence analysis
2.1. Gene finding
2.2. non-coding sequences
2.3. regulatory sequences finding
2.4. orthologous and paralogous seuqences
2.5. evolution
Layer 3. Protein sequence analysis [layer partly equivalent to layer 2]
3.1. homology detection
3.2. alignment
3.3. functional annotation
3.4. cellular localization
Layer 4. From linear sequence to three-dimensional shapes: protein folding
4.1. unfolded chain and conformational space
4.2. folding intermediates
4.3. driving forces of folding
4.4. models for protein folding
4.5. discriminating structures
4.6. misfolding
4.7. unfolding
4.8. conformational ambiguity
4.9. helpers of folding (chaperones)
Layer 5. Predicting functional structures (DNA - RNA - proteins - lipids - carbohydrates)
5.1. Homology modeling in proteins
5.2. ab initio
5.3. templates
5.4. partial information
- overall architecture
- binding pocket
- protein backbone
Layer 6. Molecular interactions
6.1. Protein-ligand
6.2. Protein-protein
6.3. Protein-DNA
6.4. Protein-RNA
6.5. Protein-lipid
6.6. Protein-carbohydrate
Layer 7. Metabolic and regulatory networks
General challenges and applications:
1. Linking variety of databases
2. Linking the different layers
3. Interpretation of experimental data
4. Design of new experiments
5. Drug screening
6. Genetic variability in individuals and in different strains (host and pathogen)