Back to "Biological Language Modeling Seminar Topics"
Back to "Introduction to Protein Protein Interactions"
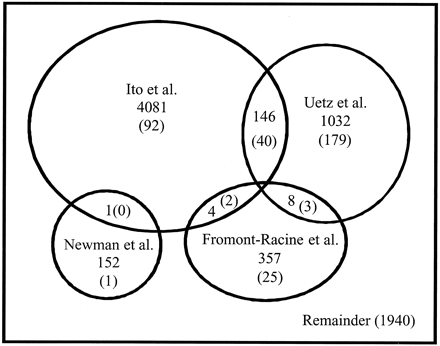
Overlap in Protein-Protein Interactions Datasets
Yeast-2-hybrid datasets:

from Deane et al 2002. All data are yeast-2-hybrid datasets.
 EPR, corresponds to
the fraction of the true positives in the experimental dataset.
Example: The
EPR, corresponds to
the fraction of the true positives in the experimental dataset.
Example: The  EPR is
calculated as 31 ± 3% (Table II) for the GY2H data,
suggesting that
EPR is
calculated as 31 ± 3% (Table II) for the GY2H data,
suggesting that  70% of the reported pairs
in this set are, in fact, false positives.
70% of the reported pairs
in this set are, in fact, false positives.
TABLE II EPR index
EPR index,  EPR,
calculated for several subsets of DIP-YEAST (see "Results" and
"Experimental Procedures" for details) using INT and RND1 subsets as
representative for the interacting and noninteracting protein populations,
respectively, is shown. The values of
EPR,
calculated for several subsets of DIP-YEAST (see "Results" and
"Experimental Procedures" for details) using INT and RND1 subsets as
representative for the interacting and noninteracting protein populations,
respectively, is shown. The values of  2
and N, the number of degrees of freedom, are given.
2
and N, the number of degrees of freedom, are given.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Mass Spec Datasets:
Gavin et al.:
7% overlap with yeast-2-hybrid datasets
56% overlap with YPD database (known complexes)
[yeast-2-hybrid has 10% overlap]
25% of all proteins are covered in Gavin study, while 95% of all proteins are covered in Yeast-2-hybrid system
Ho et al:
Table 1. in HoNature2002.pdf