Back to "Biological Language Modeling Seminar Topics"
Back to "Introduction to Protein-Protein Interactions"
Experimental identification of protein-protein interaction networks by yeast two hybrid system
Principle (http://www.devbio.com/chap05/link0518.shtml)
Let us say that you wanted to know if any particular proteins bound to protein X. Such proteins can be found by the yeast two-hybrid system (1, 2). The two-hybrid system allows in vivo detection of protein-protein interactions as well as the analysis of the affinity of these interactions.
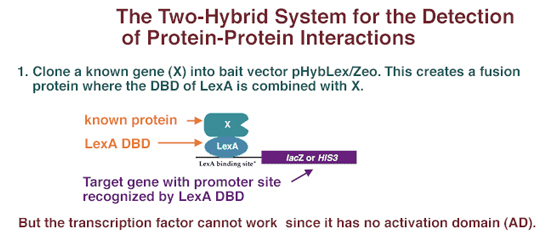
Two-hybrid technology exploits the fact that transcriptional activators are modular in nature. Two physically distinct functional domains are necessary to get transcription: (1) a DNA binding domain (DBD) that binds to the DNA of the promoter and (2) an activation domain (AD) that binds to the basal transcription apparatus and activates transcription. In the yeast two-hybrid system, the known gene encoding X, is cloned into the "bait" vector. In this way, the gene for X is placed into a plasmid next to the gene encoding a DNA-binding domain from some transcription factor. For instance, if the gene for X is cloned into the pHybLex/Zeo vector, X would be expressed as a fusion protein containing bacterially-derived LexA DBD.
Separately, a second gene (or a library of cDNAs encoding potential interactors), Y, is cloned in frame adjacent to an activation domain of a different transcription factor. For instance, it could be inserted next to the DNA encoding the B42 activation domain (AD) in a "prey" vector such as pYESTrp2.
Thus, in one strain of yeast, a known protein X is fused to the DNA binding domain of a transcription factor; and in another strain, unknown proteins are fused to the activation domain of another transcription factor. If the one of the unknown proteins combines with X, it will bring the AD over to the DBD, and transcription will be activated. So the plasmids containing the "bait" (known protein/DBD) and "prey" (unknown protein/AD) are then placed into a yeast strain, where a marker gene has a promoter containing the sequence bound by the bait protein DBD. However, in order to form a working transcription factor, it needs the AD provided by the "prey." The only way that it can get this activation domain is if the known protein X combines with some unknown protein Y that is carrying the AD. If the X and Y proteins interact, the B42 AD is brought into proximity of the LexA DBD and transcription of the reporter gene is activated (Figure 1). The activation of the reporter gene can be screened by enzyme activity, light release, or cell growth, depending on the type of reporter gene activated.

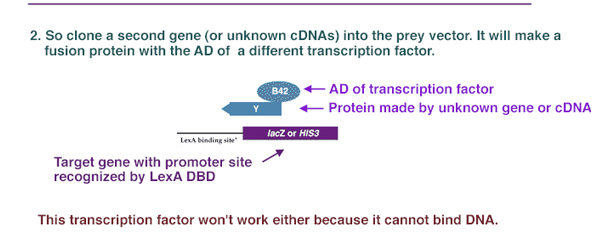
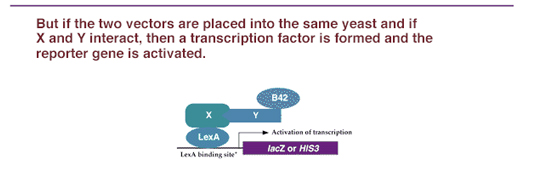
Figure taken from http://www.devbio.com/chap05/link0518.shtml