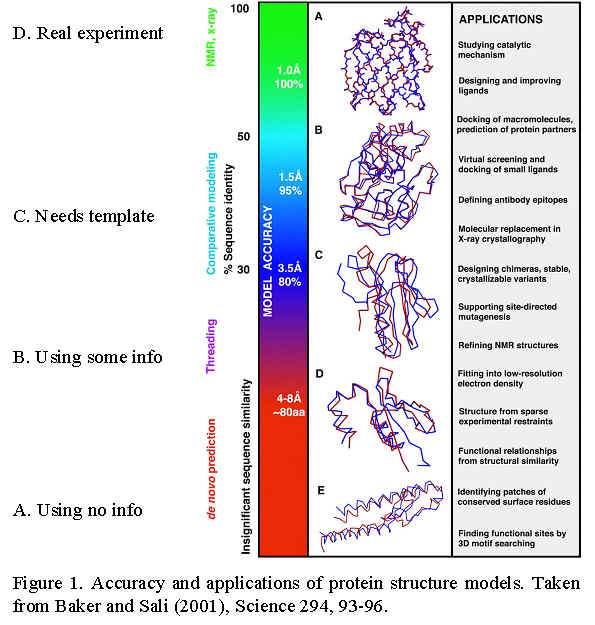
Overall survey of structure prediction: Baker and Sali 2001 Science Paper
A. When no information but sequence and physical principles are used
= ab initio structure prediction
B. When other information is used (Survey of "ab initio" methods that use pdb information and their relation to protein folding)
"fold recognition":
requires a method for evaluating the compatibility of a given sequence with a given folding pattern
B0. 3D profiles
B1. Rosetta: conformations from short segments in pdb
B2. Including experimental structural constraints
B3. Threading (=sequence-structure alignment),
B4. Inverse threading and folding experiments Reference Ivet
B4a. using short-range information
B4b. using short- and long-range information
B4. Predicting structural class only Reference Ivet
B5. Predicting active site only?
B6. Predicting protein-protein interaction sites?
B7. Predicting surface shape?
C. When a template with known structure must be available
D. Modeling structures based on experimental data
Both NMR and X-ray underdetermine the protein structure. To solve a structure one must minimize a combination of the deviation from the experimental data and the conformational energy:
D1. NMR (set of constraints on distances and angles)
D2. X-ray crystallography (Fourier transform of the electron density)