CARNEGIE MELLON UNIVERSITY
15-826 - Multimedia databases and data mining
Spring 2007
Homework 3 - Due: Tuesday April 24, 1:30pm,
in class.
Important:
- Due date: Tuesday April 24, 1:30pm, hard copy in class and soft
copy e-mailed to the TA
in a single e-mail.
- For all questions please BOTH:
- e-mail the TA
a soft-copy of all code, output (including images of plots) and auxiliary scripts tar'ed up in a
single file. Please name this file <your_andrew_id>_hw3.tar
- hand in a hard-copy of all code, output and auxiliary scripts
- Please turn in a typed report - handwritten material may not be graded,
at the grader's discretion.
- All homeworks including this one are to be done INDIVIDUALLY
- For all plots, in addition to a hardcopy printout, please submit an electronic version in postscript, pdf, gif, jpeg, or png format.
- Wherever numerical answers are asked for please provide up to 3 significant digits of accuracy.
- Expected effort for this homework:
- Q1: 1 hour
- Q2: 1 hour
- Q3: 1 hour
- Q4: 2 hours
- Q5: 1 hour
- Q6: 2 hours
- Q7: 4 hours
Q1: 1-D DFT [10 pts]
Apply DFT to following datasets.
For each dataset hand in:
- [1 pt] The time-plot (value vs time) of the original
dataset,
- [5 pt] The amplitude-frequency ("amplitude spectrum") plot
for DFT.
- [2 pt] What can you say given these plots, e.g. how many strong frequencies are there?
- [1 pt] Give the sum of squares of the time values.
- [1 pt] Give the sum of squares of the amplitudes.
Q2: 1-D DWT [10 pts]
Apply DWT using Haar
wavelets (code available here) and DFT to this simulated heart beat.
What to hand in:
- [1 pt] The time-plot of the original
dataset,
- [1 pt] The amplitude-frequency ("amplitude spectrum") plot
for DFT.
- [1 pt] What can you say given these plots?
- [2 pt] Plot the scalogram for DWT as a set of histograms (1 per scale). Do
the full level decomposition of the signals (
you might consider using
xwpl
(link
seems to be broken), so feel
free to use anything you wish, like matlab's wavelet toolbox, or see BBoard for
alternatives). The histograms should look like the examples starting on
slide #100 in in these slides, that is:
for (scale in 0 to 9){
give the 2^scale coefficients as a histogram over time
}
also give the smooth coefficient
- [1 pt] What can you say given these plots? Any dominant frequencies? Any spikes?
- [1 pt] The
time plot of the reconstructed version using only the
top 5 DFT coefficients
Specifically, pick the 5 best complex numbers of the DFT, that can help
you reconstruct the original signal. For the upcoming comparison with
DWT, we shall use the concept of 'degrees of freedom' DOF,
counted as follows:
- every complex number Xf contributes 2 towards the degrees of freedom
- except for X0 and X512, which by theorem have zero imaginary part
- Also, when you choose Xf , you can (and should) use XN-f, for free.
For the top 5 DFT coefficients, the degrees of freedom DOF would be approximately DOF=10 (or a bit less).
- [1 pt] Give the
root mean squared error (RMSE) between this reconstructed version and
the original, where RMSE is defined as the square root of the average
squared differences between the true values, y, and the reconstructed values, y^:

- [1 pt] Give the
time plot of the reconstructed version using only the top DOF DWT coefficients,
where DOF is the degrees of freedom for DFT, defined above. (Notice
that for DWT all coefficients are real, and thus each coefficient
corresponds to 1 degree of freedom).
- [1 pt] The
root mean squared error (RMSE) between this reconstructed version and the original
Q3: 2D DFT and DWT [10 pts]
Download this 2D black and white image of
16*16 pixels,
each with intensity 0 or 1. The origin of the image (x=1, y=1) is in
the top left corner (like a matrix). The format for the data is "x y
value" where value is the black-white intensity (0 = black, 1 = white).
Hand in the following:
- [2 pt] The 2-d image of the
dataset,
- [2 pt] Draw the 2-d grey-scale image of the reconstructed version
using only the top 3 DFT coefficients as defined in Q2 above. The degrees of freedom (DOF) should be about 6.
- [2 pt] What is the
root mean squared error (RMSE) between this reconstructed version and
the original, where RMSE is defined as before, but this time averaging
over all pixels (ie, all values of i and j):

- [2 pt] Draw the reconstructed
version using only the top DOF DWT coefficients, as defined in Q2 above. The idea, again, is to make it a fair comparison between DFT and DWT. (Use Haar
wavelets:
DWT each row,
then
each column).
- [2 pt] What is the
root mean squared error (RMSE) between this reconstructed version and the original?
Q4: Iterated Function Systems [15 pts]
- Dragon Curve
Write code to generate and plot the following curve for any order n:
The picture below (figure 1) shows four iterations of the dragon curve,
from order 0 on the left, to order 3 on the right. The white lines in
each image are the current state of the curve. At each iteration, each
line segment is split into two line segments by forming a 45-45-90
degree triangle (right angles shown as yellow boxes) with the original
line segment (in red) as the hypotenuse. The orientation of this new
triangle oscillates, so in the order-2 curve, the lower triangle is
formed to the right of the red line, while the upper triangle is formed
to the left of the red line.
Order 0 Order
1 Order
2 Order 3
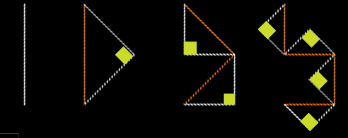
(Figure 1)
Please submit:
- [3 pts] The plot of the curve at order 6 (your code
should generate a set of 2-d line segments, which you can plot using
your favorite utility, eg, gnuplot, xgraph, etc)
- [2 pts] The fractal dimension of the curve in the limit (as n goes to infinity)
- [2 pts] Your code
- X-shape
Get the IFS code here. Now use it to plot the
following X-shape. The picture below (figure 2) shows one iteration of
the X-shape. The square is subdivided into nine sub-squares, with the
four corners and center square filled, and the rest left empty. For the
next iteration, each filled sub-square is further subdivided and filled
according to the same pattern.

(Figure 2)
Please submit:
- [2 pts] The plot of 1,000 points that you generated using Barnsley's IFS algorithm (in the ifs code)
- [2 pts] The fractal dimension of the x-shape
- [2 pts] The correlation integral (distance vs # pairs) of your 1,000 points (you can use the code here)
- [2 pts] Your code (ie, the parts of the IFS code that you modified. Eg, input.parms)
Q5: SVD [10 pts]
Consider the following Document by Term matrix:
DocTerm
The file is in csv (comma-separated-values) format with windows end of line
termination. In this file each row denotes a document, and the columns denote terms. An
entry (i,j) denotes the number of occurrences of the jth term in the
ith document. Based on their occurrence in the documents, the terms
can be clustered as belonging to a particular Topic. Run SVD on the
dataset to determine the number of topics present. You can use any standard
software (e.g., Matlab).
Hand in:
- [3 pts] The SVD components (the U, lambda, V matrices, specifying which is which).
- [1 pts] Your estimate of the number of topics for the DocTerm dataset.
- [6 pts] For every
topic, give a sorted list of documents (specified by row-id) that
participate in this topic, along with their weight in the topic. For
example:
Topic A:
--------
doc 1, weight .9
doc 2, weight .2
doc 3, weight .8
Topic B:
------
doc 1, weight .1
doc 2, weight .8
doc 3, weight .2
Q6: 3-D SVD [15 pts]
Recall the multivariate Guassian distribution in N dimensions:

where sigma is an NxN covariance matrix, and mu is a length-N mean vector.
This dataset contains 10,000 points constructed in the following manner:
-
Points are sampled from a three dimensional (N=3) spherical (Sigma is now the 3x3 identity matrix) Gaussian centered at the origin (mu is now the vector: <0, 0, 0>).
- The first coordinate of the sampled point is multiplied by a scalar a
- The second coordinate of the sampled point is multiplied by a scalar b
- The third coordinate of the sampled point is multiplied by a scalar c
- These warped points are all finally rotated and translated by
some unknown affine transformation (which is the same for all points).
[15 pts] Given this data, recover a, b and c.
HINT: Inspect the 2-d scatterplots.
Q7: Regression and Recursive Least Squares [30 pts]
Implement the method of Recursive Least Squares (RLS), with forgetting, from the paper by
[Yi et al, ICDE 2000] (pdf, ps.gz), or from [Chen & Roussopoulos, SIGMOD 94] (pdf, ps.gz).
- Language:
You may choose among C/C++, perl, python, Java, R, Matlab. However,
your algorithm must be
incremental, processing one row of data at a time.
- Input
format: n+1 numbers per line, blank separated; the last
column is the dependent variable
<x1> <x2> ... <y>
Output
format: n regression coefficients, w1 ... wn,
so that
<last-column> ~ w1 * <column-1> + ... wn
* <column-n>
For example, you could use this python code
which (a) needs "Numerical Python", and (b) has no forgetting factor.
You can "tar xvf" and then "make" it.
Note: The code is provided to mainly show you the expected
formats of
the input and output. Make sure you test it thoroughly, or rewrite it
from scratch. If you choose to rewrite it make sure that your RLS algorithm is incremental, processing one line at a time.
Apply the recursive least squares (RLS) algorithm to this simulated, fictitious dataset.
The data contains 1024 rows of three blank-separated values. Each row
represents the weekly measurement of the same patient over 20 years.
The format is:
<calories consumed> <hours of exercise performed> <cholesterol>
The first two variables are the independent variables, x1 and x2, while
the third variable (cholesterol) is the dependent variable, y. We
believe the data is generated by some linear function, whose parameters
w1 and w2 we are trying to estimate:
y = w1*x1 + w2*x2 + noise
At some point in time, the patient began taking a medicine to lower her
cholesterol. We would like to identify a) when the patient began taking
the medicine and b) what effect (if any) this medicine had on the
relationship between the patient's calories, exercise and cholesterol.
We also want to investigate how much of a difference running ordinary
least squares (OLS, read more here or see property C1 in slide #17 in these slides, which is equivalent to equation 4 in this paper) and RLS makes on the results. At every week we estimate w1(t) and w2(t) that best fit the data up to and including week t.
What to hand in:
- Plot both the time-series of how w1(t) and w2(t) change over time, and the 2-d scatter plot of <w1(t), w2(t)> over time t, as estimated by:
- [4 pts] RLS with no forgetting (ie, lambda = 1)
- [4 pts] RLS with the forgetting factor lambda = 0.95
- [4 pts] OLS with no forgetting (ie, lambda = 1)
- [4 pts] OLS with the forgetting factor lambda = 0.95
- [2 pts] Based on these plots, when (which week) did the patient begin taking her medication?
- [2 pts] Give the running times (wall clock) of each of the
four experiments, and the specifications of the machine you ran it on
(eg, win xp, 256 mb ram, intel dual core 3 ghz)
- [10 pts] Submit your code for both RLS and OLS
Last modified: April 19, 2007