

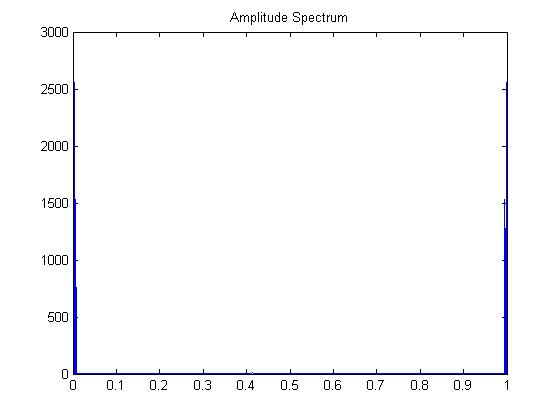
One strong frequency (but notice the symmetry/reflection over
.5)
520.7175
5.3321e+005
C.f. Parseval's
theorem:

"where X[k] is the DFT of x[n], both of length N."
Note that Matlab does not do the normalization by N for you, so you
must normalize the numbers Matlab gives you in order to see
Parseval's equivalence:
sum of squares of the time values = (1/# samples) * sum of squares
of the amplitudes
in this case: 520.7175 = 1/1024 * 5.3321e+005

One strong frequency, and another strong, but weaker one.
1.7409e+004
1.7827e+007


No strong frequency stands out -- just noise
350.7012
3.5912e+005

























