Click here for link to latest DREM website
Supporting Website for:
Reconstructing dynamic regulatory maps
Jason Ernst, Oded Vainas, Christopher T. Harbison, Itamar
Simon,
and Ziv
Bar-Joseph
Nature-EMBO Molecular Systems Biology, 3:74, 2007
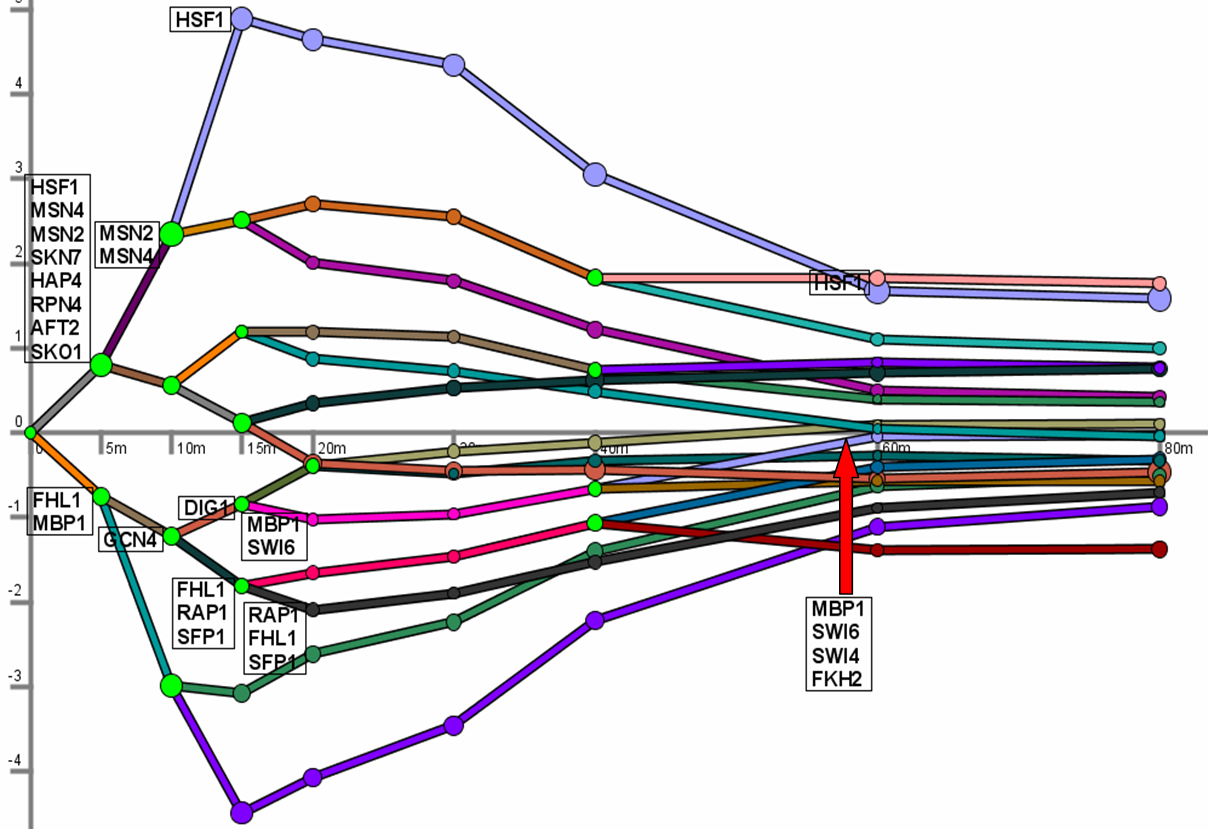
Abstract
Even simple organisms have the ability to respond to internal and external stimuli. This response is carried out by a dynamic network of protein-DNA interactions that allows the specific regulation of genes needed for the response. We have developed a novel computational method that uses an input-output hidden Markov model to model these regulatory networks while taking into account their dynamic nature. Our method works by identifying bifurcation points, places in the time series where the expression of a subset of genes diverges from the rest of the genes. These points are annotated with the transcription factors regulating these transitions resulting in a unified temporal map. Applying our method to study yeast response to stress, we derive dynamic models that are able to recover many of the known aspects of these responses. Predictions made by our method have been experimentally validated leading to new roles for Ino4 and Gcn4 in controlling yeast response to stress. The temporal cascade of factors reveals common pathways and highlights differences between master and secondary factors in the utilization of network motifs and in condition-specific regulation.
New Experimental Data
Gcn4 ChIP-chip in MMS: gcn4bindingpvals.txt, Gcn4 in EBI (E-MEXP-905)
Ino4 ChIP-chip in AA: ino4bindingpvals.txt, Ino4 in EBI (E-MEXP-906)
Software - Dynamic Regulatory Events Miner (DREM)
Funding was supported in part by NIH grant NO1 AI-5001 and NSF CAREER award 0448453 to ZBJ