|
|
Performance
Comparison with Only Top 20 detail features
- For the detailed feature type,
to find how the most important features for each gold standard set by
RF-Gini Importance perform
- To see the
performance dropping when only use the top N important features. Here we
use N = 20.
- Derived performance for each
gold standard based on each’ top 20 features within the detail style
- Precision vs. recall curves
comparisons: through Random Forest (RF) - 200 tress: ( Top20 features vs.
Full-Detail 162 features )
- è The top 20 detail features
did a pretty good job compared to the full 162 detail feature set
1. Co-Complex Protein Pair Prediction Summary

- The Top 20 features ordering in
the training/test sets (MIPS)
- 3 4 5 8 9 13 14 16 19 20 23 39
48 49 61 76 85 100 101 105
- The Top 20 features for the
MIPS (co-complex task):
|
|
Feature Index |
Description |
Note: |
The RF-Imp + Top 20 Feature Ranked list |
|
1 |
3 |
Sporulation |
Gene exp |
0.0059 61 0.0061 85 0.0068 100 0.0070 105 0.0090 14 0.0095 23 0.0100 48 0.0139 20 0.0228 4 0.0241 3 0.0385 9 0.0430 5 0.0441 13 0.0569 16 0.0715 8 0.0731 101 0.0778 19 0.0965 39 0.1182 76 0.1637 49 |
|
2 |
4 |
fermentation to respiration / cell wall |
Gene exp |
|
|
3 |
5 |
Eisen1998 stress |
Gene exp |
|
|
4 |
8 |
Jelinsky2000 alkylating agents |
Gene exp |
|
|
5 |
9 |
Lyons2000 zinc |
Gene exp |
|
|
6 |
13 |
Shamji2000 |
Gene exp |
|
|
7 |
14 |
Travers2000 |
Gene exp |
|
|
8 |
16 |
Gasch2001 |
Gene exp |
|
|
9 |
19 |
Gasch2000 stress |
Gene exp |
|
|
10 |
20 |
cell
cycles |
Gene exp |
|
|
11 |
23 |
transcription
regulator activity |
Go Func |
|
|
12 |
39 |
structural
molecule activity |
Go Func |
|
|
13 |
48 |
protein
catabolism |
Go Proc |
|
|
14 |
49 |
protein
biosynthesis |
Go Proc |
|
|
15 |
61 |
Transcription |
Go Proc |
|
|
16 |
76 |
Ribosome |
Go Comp |
|
|
17 |
85 |
mitochondrial membrane |
Go Comp |
|
|
18 |
100 |
HMS_PCI Mass |
|
|
|
19 |
101 |
TAP Mass |
|
|
|
20 |
105 |
Sequence Similarity |
|
2. Direct Protein-Protein Interaction
Prediction Summary
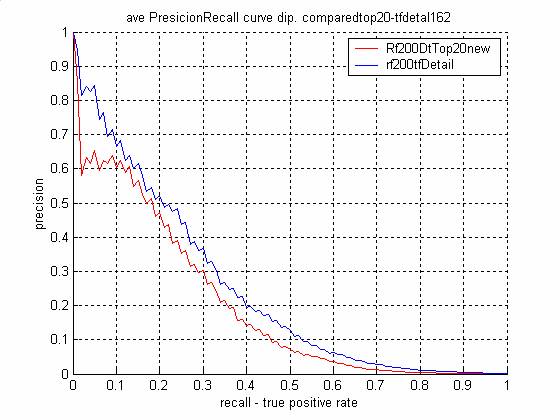
- The Top 20 features ordering in
the training/test sets (DIPS)
- 3 5 8 20 23 30 48 61 67 74 83
100 101 102 103 105 109 110 157 162
- The Top 20 features for
DIPS physical binding task
|
|
Feature Index |
Description |
Note: |
The RF-Imp + Top 20 Feature Ranked list |
|
1 |
3 |
sporulation |
Gen Exp |
0.0130 109 0.0140 162 0.0140 30 0.0141 74 0.0153 48 0.0171 23 0.0172 61 0.0189 157 0.0200 3 0.0209 103 0.0234 20 0.0261 83 0.0267 8 0.0306 105 0.0336 5 0.0352 110 0.0372 67 0.0459 100 0.0611 102 0.1035 101 |
|
2 |
5 |
Eisen1998 stress |
Gen Exp |
|
|
3 |
8 |
Jelinsky2000 alkylating agents |
Gen Exp |
|
|
4 |
20 |
cell
cycle ( 76 col ) |
Gen Exp |
|
|
5 |
23 |
transcription
regulator activity |
Go Func |
|
|
6 |
30 |
RNA binding |
Go Func |
|
|
7 |
48 |
protein catabolism |
Go Proc |
|
|
8 |
61 |
transcription |
Go Proc |
|
|
9 |
67 |
nucleus |
Go Proc |
|
|
10 |
74 |
Golgi
apparatus |
Go Proc |
|
|
11 |
83 |
vesicle-mediated transport |
Go Comp |
|
|
12 |
100 |
HMS_PCI Mass |
HMS_PCI Mass |
|
|
13 |
101 |
TAP Mass |
TAP Mass |
|
|
14 |
102 |
Y2H |
Y2H |
|
|
15 |
103 |
Synthetic Lethal |
Synthetic Lethal |
|
|
16 |
105 |
Celegan homology PPI |
Sequence Similarity |
|
|
17 |
109 |
celegan |
Homology |
|
|
18 |
110 |
Domain-Domain Interaction |
Domain-Domain Interaction |
|
|
19 |
157 |
Cell cycle defects |
MIPS protein phenotype |
|
|
20 |
162 |
Nucleic acid metabolism defects |
MIPS protein phenotype |
3. Co-pathway Protein Pair Prediction Summary
- The Top 20 features ordering in
the training/test sets (KEGG04)
- 3 4 5 8 9 13 14 16 19 20 24 28
39 41 49 51 76 98 104 157
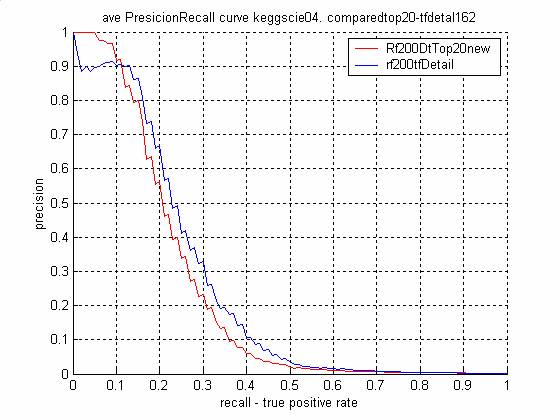
- The Top 20 features for keGG04
|
|
Feature Index |
Description |
Note: |
The RF-Imp + Top 20 Feature Ranked list |
|
1 |
3 |
sporulation |
Gen Exp |
0.0118 41 0.0127 98 0.0142 24 0.0142 28 0.0147 14 0.0170 20 0.0204 3 0.0207 104 0.0254 13 0.0255 4 0.0310 157 0.0356 51 0.0474 9 0.0511 39 0.0571 8 0.0584 16 0.0726 19 0.0729 5 0.0757 76 0.0867 49 |
|
2 |
4 |
fermentation to respiration / cell wall |
Gen Exp |
|
|
3 |
5 |
Eisen1998 stress |
Gen Exp |
|
|
4 |
8 |
Jelinsky2000 alkylating agents |
Gen Exp |
|
|
5 |
9 |
Lyons2000 zinc |
Gen Exp |
|
|
6 |
13 |
Shamji2000 |
Gen Exp |
|
|
7 |
14 |
Travers2000 |
Gen Exp |
|
|
8 |
16 |
Gasch2001 |
Gen Exp |
|
|
9 |
19 |
Gasch2000 stress |
Gen Exp |
|
|
10 |
20 |
cell cycle |
Gen Exp |
|
|
11 |
24 |
transferase activity |
Go Func |
|
|
12 |
28 |
oxidoreductase activity |
Go Func |
|
|
13 |
39 |
structural molecule activity |
Go Func |
|
|
14 |
41 |
nucleotidyltransferase activity |
Go Func |
|
|
15 |
49 |
protein biosynthesis |
Go Proc |
|
|
16 |
51 |
carbohydrate metabolism |
Go Proc |
|
|
17 |
76 |
ribosome |
Go Comp |
|
|
18 |
98 |
Prot Exp |
Prot Exp |
|
|
19 |
104 |
Gene Neigh/Fusion/Cocur |
Gene Neigh/Fusion/Cocur |
|
|
20 |
157 |
Cell cycle defects |
MIPS phenotype |