Final Report - PDF
Overview
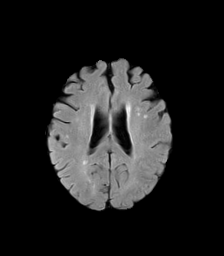
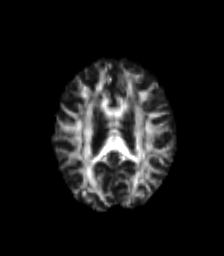
The goal of this project is to design a MRI(magnetic resonance imaging) mode invariant feature point descriptor. Different MRI modes (T1, T2, and diffusion fractional anisotropy imaging) each provide differnet sets of information about the underlying anatomical structure. However, there is always an inevitable overlap in information between these image types. I intend to investigate whether this information overlap is sufficient to allow the matching of interest points between images taken of the human brain with different MRI modalities. These interest point correspondences would be useful for non-rigid multimodal registrations, as well as feature recognition. Below, we can see images of the three MRI modes I will be investigating. Bright patches in one image type can appear much darker in others, complicating the uses of standard interest point descriptors.
 |
 |
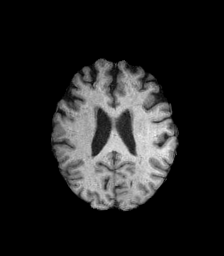 |
| T2 | Fractional Anisotropy | T1 |
Previous work
Multimodal interest point based registration has been done before, however the matching was not descriptor based. Wong and Bishop [1] use prior knowledge from successful alignments (in the form of stored matched image patches) in order to predict geometric transformations of the current query images. Previous work has also been done with SURF based interest point matching, but not in a multimodal setting [2]. The combination of interst point matching with multimodal feature points provides a novel aspect to this project.
Methods and Data
This project will incorporate the methods in [3] to help design a descriptor capable of matching interest points across MRI modalities. I intend to implement at least a portion of Brown et al's descriptor learning framework, using a dataset consisting of sets of T1, T2, and fractional anisotropy images that have been aligned so that ground truth correspondences can be easily obtained for deteced interest points. While the dataset that I have access to is in fact a full 3D volume set (~20 subjects), I will likely only address 2D points for this project.
 |
Results
ROC curves for the interest point descriptors were constructed for both the standard SIFT implementation and for the custom pipelines.
Standard SIFT descriptor ROCs
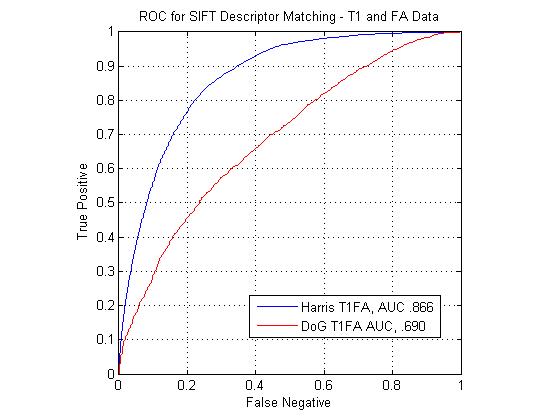
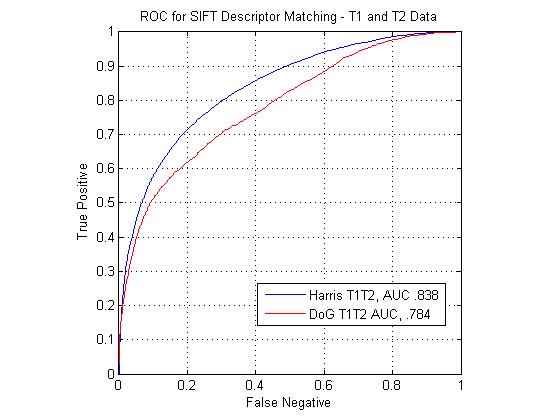
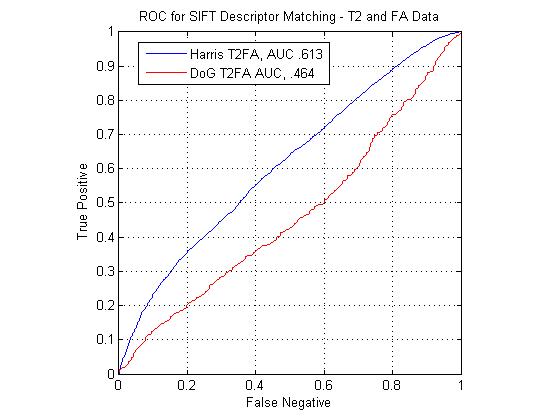
The following figure shows ROC curves for matching points using the standard SIFT descriptor computed at Harris corners and DoG interest points. The SIFT descriptors extracted at Harris corners can be seen to outperform the DoG descriptors for all modality pairs. The figure shows that the T1-FA match pair performs the best with the standard SIFT descriptor. The T2-FA performs the worst, barely above chance. This is understandable, given how few true matches interest point matches could be found between the two image types.
 |
 |
 |
 |
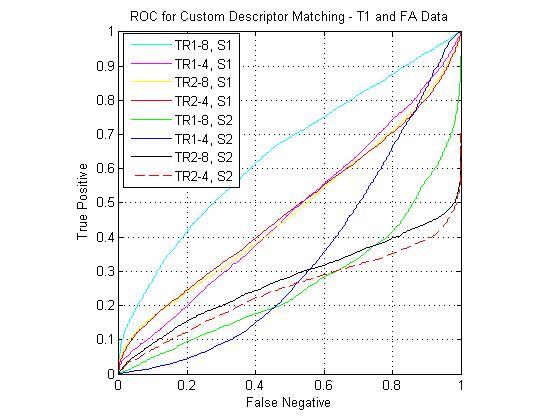
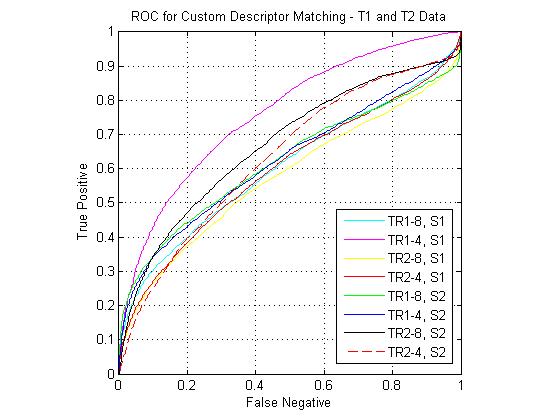
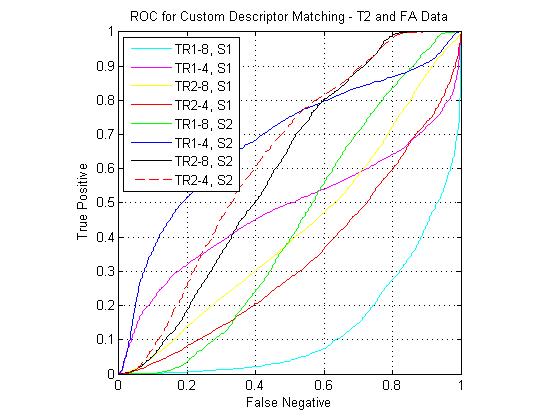
Custom descriptor ROCs
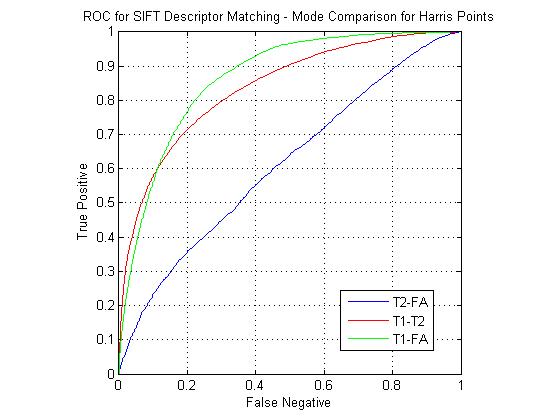
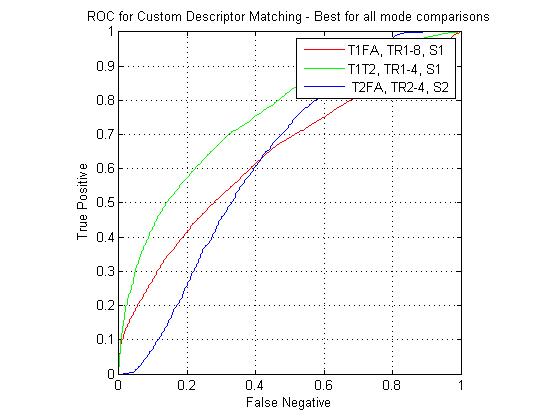
The following figure shows ROC curves for matching using the eight custom pipelines for all three match combinations. The optimized custom pipelines gave poor performance compared to the standard SIFT descriptor, including the TR1-8, S1 pipeline, which mirrors the SIFT design. I have been continuing to investigate the source of this large error. However, I still feel that the custom pipeline results are valid for comparison to each other. For instance, the comparative performance of the SIFT clone pipeline (T1-FA > T1-T2 > T2-FA) was the same as that of the standard SIFT. For the T1-FA pairing, the best pipeline was the SIFT clone, which is to be expected given the good performance given by the standard SIFT. For T1-T2 the best pipeline was the TR1-4, S1 combination (i.e. coarsely binned gradient version of SIFT). For T2-FA, the results were much more varied, however the best performers used the S2 block (DAISY style spatial pooling).
 |
 |
 |
 |
Conclusions
The overall results were underwhelming due to the as yet unknown errors in the custom descriptor pipelines. However, the one interesting result was the performance of the DAISY based descriptors for matching between the T2 and FA interest points. Future work for this project will include designing the custom descriptors using image patches extracted at arbitrary match points (i.e. not restricting to point that are discovered by the interest point detector). A dense formulation is also being considered.
References
- Wong, A.; Bishop, W.; , "Indirect Knowledge-Based Approach to Non-Rigid Multi-Modal Registration of Medical Images," Electrical and Computer Engineering, 2007. CCECE 2007. Canadian Conference on , vol., no., pp.1175-1178, 22-26 April 2007
- Wang, A.; Zhe Wang; Dan Lv; Zhizhen Fang; , "Research on a novel non-rigid registration for medical image based on SURF and APSO," Image and Signal Processing (CISP), 2010 3rd International Congress on , vol.6, no., pp.2628-2633, 16-18 Oct. 2010
- Brown, M.; Gang Hua; Winder, S.; , "Discriminative Learning of Local Image Descriptors," Pattern Analysis and Machine Intelligence, IEEE Transactions on , vol.33, no.1, pp.43-57, Jan. 2011 doi: 10.1109/TPAMI.2010.54