
Back to "Biological Language Modeling Seminar Topics"
Back to "Protein structure prediction"
Description of CATH
taken directly from http://wserv1.dl.ac.uk/CCP/CCP11/newsletter/vol2_3/orengo/
1 Biomolecular Structure and Modelling Unit, Department of
Biochemistry and Molecular Biology, Gower St, London WC1E 6BT.
2 Department of Crystallography, Birkbeck College, Malet Street,
London WC1E 7HX.
The CATH database of protein domain structures classifies structures according to their (C)lass, (A)rchitecture, (T)opology or fold and (H)omologous family (http://www.biochem.ucl.ac.uk/bsm/cath). Although the protocol used is mostly automatic, manual inspection is used to check assignments at some critical stages, such as the detection of very distantly related homologues and analogues and the assignment of novel architectures. We have recently established a facility to search the database with the coordinates of a newly determined structure. The CATH server can be accessed over the World Wide Web (http://www.biochem.ucl.ac.uk/bsm/cath/server/) and mirror sites are planned to improve access.
With the huge increases in the Brookhaven Protein Databank (PDB, Abola et al. 1987) expected - some estimates suggest that there will be as many as 20,000 structures in total by the millenium, it is essential to automate classification as far as possible. In CATH, we use an automatic structure comparison method for identifying homologues or analogues, with cutoffs established by empirical trials (SSAP, Taylor & Orengo, 1989, Orengo et al. 1992). Proteins are grouped into families according to global similarities in their structures (Orengo et al. 1997).
The four major hierarchical levels in CATH are class, architecture, topology (or fold) and homologous family. There are currently three major classes recognised in CATH (mainly-alpha, mainly-beta and alpha-beta). Below class, the architecture level simply describes the orientations of the secondary structures in 3D without regard to their connectivity. We currently recognise 28 well-defined architectures and within each architecture, the topology or fold is determined by the connectivity of the secondary structures (figure 1).

Figure 1: Schematic representation of the (C)lass, (A)rchitecture and (T)opology/fold levels in the CATH database
This gives ~600 fold families in CATH and structures are further grouped into homologous families whenever there is sufficient evidence of an evolutionary relationship. Within each homologous superfamily, proteins are subgrouped according to sequence similarity, measured as percent identity (S-level (>=35%), N-level (>=95%), I-level (100%)). Analysis of CATH reveals that currently less than one quarter of new sequences (i.e. those which have less than 25% sequence identity to any structure in CATH), are found to adopt a novel fold. This suggests that we may now have representatives for many of the major folds occurring in nature.
CATH currently contains >12,000 chains from the Brookhaven Protein Databank. Only well-resolved (< 3.0A) structures are included and there are no models, or synthetic proteins. Each entry in CATH is linked to the PDBSUM database (Laskowski et al. 1997) which generates web pages showing summary information derived from the PDB file.

Figure 2: Pyramid plot showing the number of groups identified at each level in the CATH database. Characters on the lefthand-side gives the CATH levels: (C)lass; (A)rchitecture, (T)opology; (H)omologous superfamily; (S)equence family - 35% sequence identity; (N)ear-identical - 95% sequence identity; (I)dentical - 100% sequence identity; (D)omain entry.
We have recently set up a server which allows the user to submit the coordinates of a newly determined structure for automatic classification in CATH. The server also provides a list of structural neighbours and alignments are given for the 5 highest scoring matches.
The CATH Server runs on a 4-processor Origin 200, obtained by funding from the Medical Research Council. Procedures used mirror all the automatic stages in the CATH classification. Domains are first assigned, before running sequence and structure comparison programs to identify a possible fold family for each domain.
We have used the DETECTIVE program (Swindells, 1995) which is relatively good at identifying multi-domain proteins. The results from DETECTIVE are returned to the user in less than a minute and domains may be further split or merged or the boundaries may be moved using a Web form.
Identified domains are scanned against non-identical representatives from CATH using a global sequence alignment method (domhomol, Orengo et al. 1997). If a sequence match with percentage identity greater than 95% is found then we assume that the domain is nearly identical to one in CATH and a link is provided to the CATH entry for that hit. If a sequence hit is found with percentage identity greater than 30% then we assume that proteins are homologous (i.e. the C.A.T.H number will be the same) and we run structure comparisons (see below) against representatives from all the sequence families (S-level) for this homologous family (H-level).
Subsequently, the fast version of the structure comparison algorithm, SSAP (Taylor & Orengo, 1989, Orengo et al. 1992) scans representatives from all the sequence families (S-level) in those possible fold families. The normal (slow) version of SSAP is then used to scan the non-identical representatives of the S-families providing the best matches. Structural pairs returning a SSAP score greater than 80 are possible homologues. Whilst lower values of 70-80 are occasionally returned by very distant homologues as well as by analogous pairs having the same fold but no sequence or functional similarity.
Barcharts showing distributions of scores are displayed together with tables of scores and hits which are clickable links back to the appropriate CATH family and derived data (see figure 3). Finally, the SSAP structural alignment between the submitted domain and the top 5 matching structures is displayed using a graphical display package (SAS, Milburn et al. 1998) (see figure 4).

Figure 3: Diagnostic report from the CATH server, giving a
histogram of SSAP scores obtained by scanning the new structure against
representatives from CATH. A list of top scoring structural neighbours is
provided, together with associated SSAP scores, sequence identities and links to
relevant CATH families.
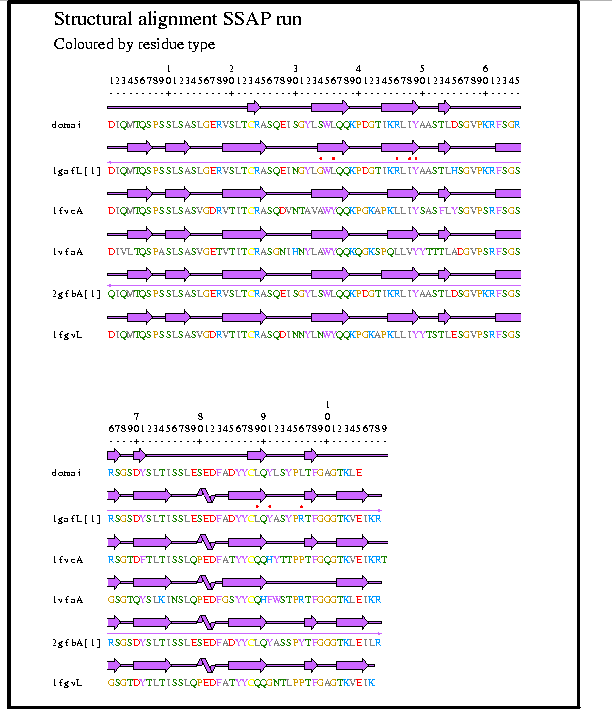
Figure 4: Diagnostic report from the CATH server showing the multiple structure alignment of the probe structure with the five highest scoring structures from the SSAP scan. The aligment plot is generated using graphical software in the SAS package (Milburn et al. 1998).
The majority of fold families in CATH (>95%) contain only homologous proteins and these often have significant sequence similarity (>25%) which is accompanied by considerable structural similarity. Recognising new relatives of these families is relatively easy as any member of the family can be expected to give a good pairwise SSAP score against a new structural relative. The remaining structural families (Superfolds, Orengo et al. 1994), which include the updown helix bundles, Immunoglobulin-like, OB, TIM barrel, Rossmann and alpha-beta plait folds, contain many diverse sequence relatives and there can be considerable structural variation across the family. Matches to proteins in these families should be considered carefully and assessed by examining both structure similarity scores and the degree of residue overlap. The TOPS server set up at EBI by David Westhead (see CCP11 Newsletter Issue 4, Vol 2.2 - 14 April 1998) can also be very helpful in checking for similarities in protein topology (http://www3.ebi.ac.uk/tops/).
Abola, E.E., Bernstein, F.C., Bryant, S.H., Koetzle, T.F. & Weng, J. (1987), Protein Databank. in Crystallographic Databases-Information Content, Software Systems, Scientific Applications (Allen, F.H., Bergerhoff, G. & Sievers, R. Eds), 107-132.
Laskowski, R.A., Hutchinson, E.G., Michie, A.D., Wallace, A.C., Jones, M.L. & Thornton, J.M. (1997) PDBsum: A Web-Based Database of Summaries and Analyses of all PDB Structures. Trends biochem Sci. 22:488-490.
Michie, A.D., Orengo, C.A. & Thornton, J.M. (1996), Analysis of Domain Structural Class using an Automated Class Assignment Protocol. J. Mol. Biol. 262:168-185.
Milburn, D. Laskowski, R.A. & Thornton, J.M (1998), SAS: Protein Eng. Submitted.
Orengo, C.A., Brown, N.P. & Orengo, C.A. (1992), Fast Structure Alignment for Protein Database Searching. Proteins. 14:139-167.
Orengo, C.A., Jones, D.T. & Thornton, J.M. (1994), Protein Superfamilies and Domain Superfolds. Nature, 372, 631-634.
Orengo, C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B. & Thornton, J.M. (1997), CATH - A Hierarchic Classification of Protein Domain Structures. Structure: 5:1093-1108.
Swindells, M.B. (1995), A Procedure for Detecting Structural Domains in Proteins. Protein Sci. 4, 103-112.
Taylor, W.R. & Orengo, C.A. (1989), Protein Structure Alignment. J. Mol.
Biol., 208:1-22.