Back to "Biological Language Modeling Seminar Topics"
Reference: www.mshri.on.ca/pawson/research1.html
Back to "Protein protein interactions"
PTB Domains
Motifs that are recognized by PTB domains:
- with secondary modification: phosphotyrosine-containing motifs,
- other domains that recognize phosphotyrosine: SH2
- commonly bind Asn-Pro-X-Tyr motifs. The PTB domains of the docking proteins Shc and IRS-1 require ligand phosphorylation on the tyrosine residue (NPXpY) for binding. More N-terminal sequences are also required for high affinity binding and conferring specificity.
Proteins with PTB domains:
- found on docking proteins such as the IRS-1 substrate of the insulin receptor.
PTB domain protein |
Binding partner and Peptide Ligand |
| Shc docking protein | TrkA Nerve Growth Factor Receptor: Ile-Ile-Asn-Pro-Gln-pTyr |
| IRS-1 docking protein | Insulin receptor: Leu-Tyr-Ala-Ser-Ser-Asn-Pro-Glu-pTyr |
| X11 neuronal protein | b-amyloid precursor
protein: Tyr-Glu-Asn-Pro-Thr-Tyr |
Versatility of PTB domains:
Although PTB domains were originally discovered through their ability to bind
phosphotyrosine in the context of an Asn-Pro-X-Tyr (NPXY) motif which forms a
b-turn, it appears that many PTB domains recognize NPXY-related peptide motifs,
but in a phospho-independent fashion. Thus PTB domains likely evolved to bind
unphosphorylated peptides, and have subsequently developed a capacity to
recognize phosphotyrosine in a few specific cases. Furthermore, an individual
PTB domain, such as those from the Numb and FRS-2 proteins, can recognize two
quite different peptide ligands.
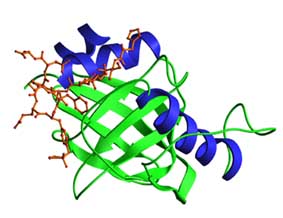
Structure
- The PTB domains of Shc and IRS-1 contain two orthogonal b-sheets and connecting loops, and have very similar folds despite their low sequence similarity. Both have a C-terminal amphipathic a-helix capping one end of the b-sandwich. The N-terminal residues of the peptide ligand form an additional anti-parallel b-strand to the second b-sheet. The figure shows the PTB domain of Shc complexed to a HIIENPQpYFS peptide.
- Phosphotyrosine binding (PTB) domains are 100-150 residue modules
- The peptide binds as a b-strand to an anti-parallel b-sheet, while the NPXpY motif makes a turn, positioning the pY for recognition by basic residues. The PTB domains of proteins such as X11, Dab, Fe65 and Numb apparently recognize NPXY or related peptide motifs, but are not dependent on ligand phosphorylation. In addition, the Numb PTB domain can bind an unrelated peptide that forms a helical turn.