Back to "Biological Language Modeling Seminar Topics"
Reference: www.mshri.on.ca/pawson/research1.html
Back to "Protein protein interactions"
SH2 Domains
Motifs that are recognized by SH2 domains:
- with secondary modification: phosphotyrosine (pY)-containing motifs,
- Examples for protein who carry this motif: activated receptors for growth factors, cytokines and antigens.
- other domains that recognize phosphotyrosine: PTB
- Conventional SH2 domains have a conserved pocket that recognizes pY, and a more variable pocket that binds 3-6 residues C-terminal to the pY. Different SH2 domains differ in their preference for the amino acids immediately following pY, which provides an element of specificity in signaling by tyrosine kinases
- The SAP SH2 domain recognizes Y as well as pY in the context of residues N and C terminal, suggesting an alternate 3-pronged model may apply in some cases.
List:
Src-like pTyr-Glu-Glu-Ile
Grb2 pTyr-X-Asn
others?
Binding constants:
Phosphopeptides of optimal sequence bind to SH2 domains with dissociation constants of ~50-500 nM.
Abundance:
the human genome is predicted to encode at least 120 SH2 domains.
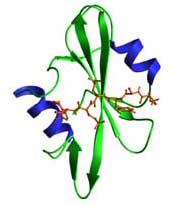
Structure of SH2 domain:
- modules of ~100 amino acids
- SH2 domains contain a central anti-parallel b-sheet surrounded by two a-helices. The phosphopeptide generally binds as an extended b-strand that lies at right angles to the SH2 b-sheet. Conserved residues contribute to the hydrophobic core or are involved in pY recognition while more variable residues contribute to specific recognition of C-terminal residues. An invariant Arg residue in the SH2 domain coordinates the phosphate oxygens of pY and is essential for high affinity phosphopeptide binding.

The figure shows the SH2 domain of v-src bound to a pYEEI peptide ligand.
Examples of proteins with SH2 domain
- SH2 domains regulate functionally diverse processes.
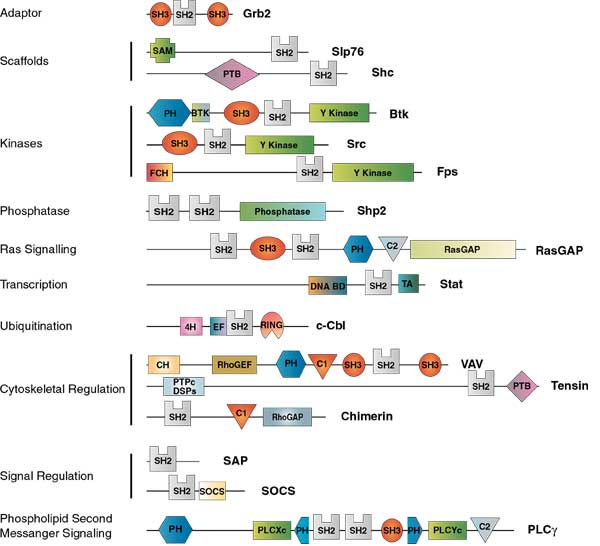
Figure: Examples of SH2 domain proteins (Reference: Pawson, T., Gish, G., and Nash, P. Trends in Cell Biology Vol.11 No.12 December 2001)
- Binding properties of SH2 domains
Conventional SH2 domains must achieve something of a balancing act.
Their affinity for an unphosphorylated site must not be too high, or
binding will not be regulated by phosphorylation. At the same time, the
SH2 domain must obtain sufficient binding energy from the recognition of
adjacent residues to allow discrimination between different
phosphorylated sites, and thus a degree of specificity. Furthermore, SH2
domains must exhibit sufficiently high off-rates for rapid and
reversible signal transduction. A structural basis for the specificity
of SH2 domain-mediated interactions has been provided by numerous
crystal and solution structures of SH2 domains bound to specific
phosphotyrosine-containing peptides (sKuriyan J, Cowburn D. Annu Rev
Biophys Biomol Struct 1997. 26, 259-288).
The SH2 domain fold, which is composed of a central anti-parallel b-sheet sandwiched between two a-helices, provides a positively charged pocket on one side of the b-sheet for binding of the ligandís phosphotyrosine moiety, and an extended surface on the other for binding to ligand residues C-terminal to the phosphotyrosine.
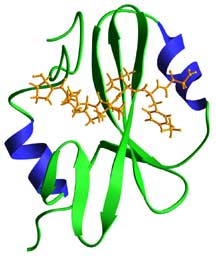
Ribbons diagram of the SH2-C domain of phospholipase-C( bound to a specific phosphotyrosine-containing peptide. The peptide (gold) is derived from the cytoplasmic domain of the PDGF receptor (DNDpYIPLPDPK) derived from PTB:2PLD (Pascal SM, Singer AU, Gish G et al. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma1 complexed with a high affinity binding peptide. Cell 1994. 77, 461-472.).
Subtle differences in the molecular architecture of this extended surface can have very significant effects on ligand-binding specificity. For example, mutation of a threonine residue in this region of the Src SH2 domain to tryptophan, found at the corresponding position of the Grb2 SH2 domain, converted ligand-binding specificity from the Src-like pTyr-Glu-Glu-Ile, to the signature Grb2 binding motif pTyr-X-Asn (Kimber MS, Nachman J, Gish G, Pawson T, Pai E. Molecular Cell 2000. 5, 1043-1049).
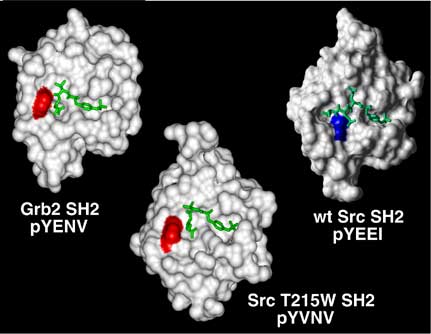
Structural analysis provides a basis for understanding SH2 domain binding specificity. Crystal structures of the Grb2 and Src SH2 domains bound to optimal phosphopeptides illustrate the different modes of ligand binding. A switch in the ligand preference of the Src SH2 domain to pTyr-Val-Asn-Val is achieved through mutation of a threonine residue, highlighted in blue on the surface of the Src SH2, to the tryptophan shown as red in the Src T215W SH2 domain. In a similar fashion, the molecular architecture of the Grb2 SH2 domain uses a tryptophan (red) to define its binding to pTyr-X-Asn ligands. (For more details see Pawson, T., Gish, G., and Nash, P. Trends in Cell Biology Vol.11 No.12 December 2001)