Back to "Biological Language Modeling Seminar Topics"
Back to "Protein structure prediction"
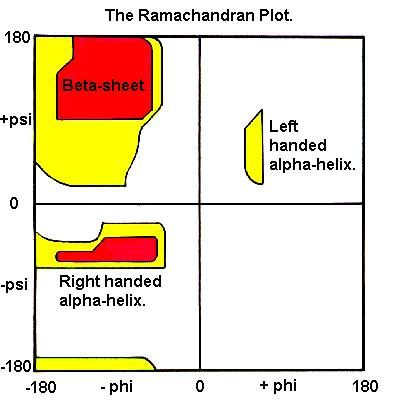
Ramachandran plot:

Figure taken from http://www.cryst.bbk.ac.uk/PPS95/course/3_geometry/rama.html on 10/13/02.
Ramachandran generated computer models of small polypeptides and systematically varied phi and psi. Atoms were treated as hard spheres with dimensions corresponding to their van der Waals radii. Phi and psi angles which cause spheres to collide (steric clashes) are disallowed conformations of the polypeptide backbone. In the figure above the white regions correspond to conformations where atoms in the polypeptide come closer than the sum of their van der Waals radi. These regions are sterically disallowed for all amino acids except glycine. Disallowed regions generally involve steric hindrance between the side chain C-beta methylene group and main chain atoms. Glycine has no side chain and therefore can adopt phi and psi angles in all four quadrants of the Ramachandran plot. The red regions correspond to conformations where there are no steric clashes, ie these are the allowed regions. The yellow regions show the allowed regions if slightly shorter van der Waals radi are used in the calculation, ie the atoms are allowed to come a little closer together.
L-amino acids cannot form extended regions of left-handed helix but occassionally individual residues adopt this conformation. These residues are usually glycine but can also be asparagine or aspartate where the side chain forms a hydrogen bond with the main chain and therefore stabilizes this otherwise unfavorable conformation. The 3(10) helix occurs close to the upper right of the alpha-helical region and is on the edge of allowed region indicating lower stability.