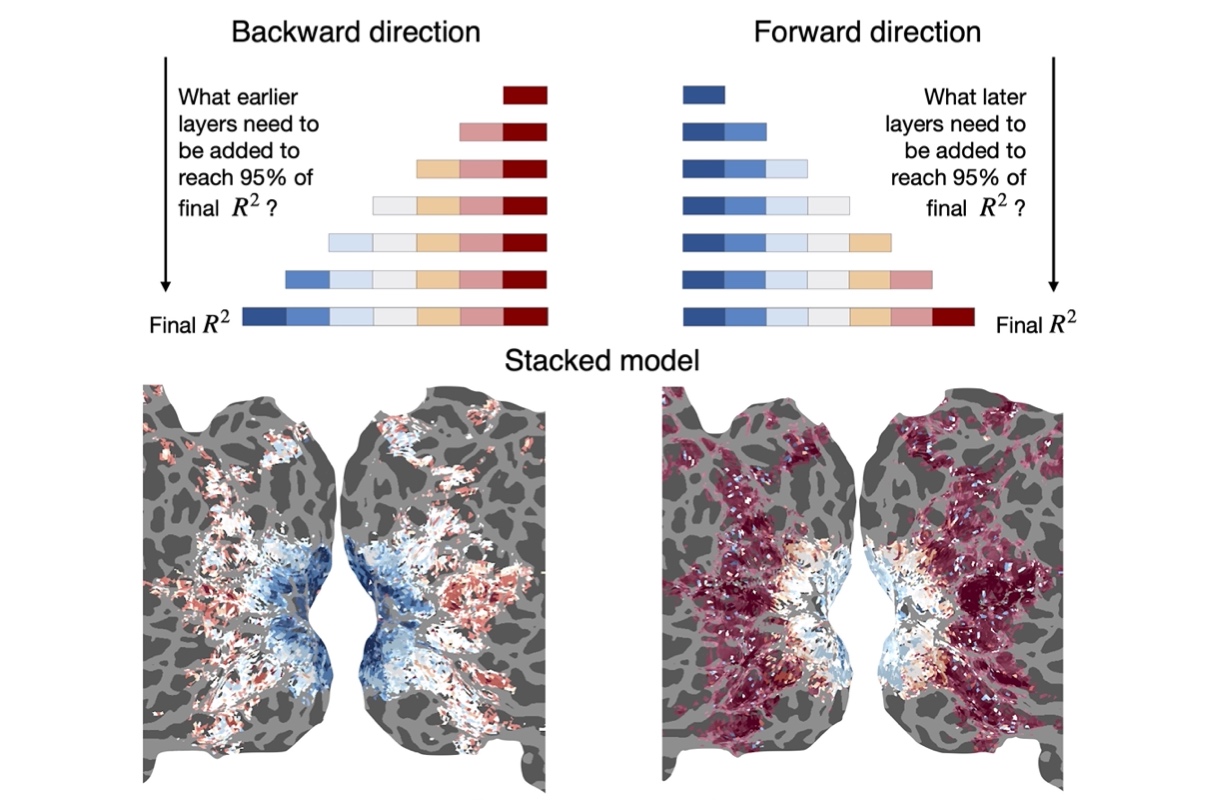
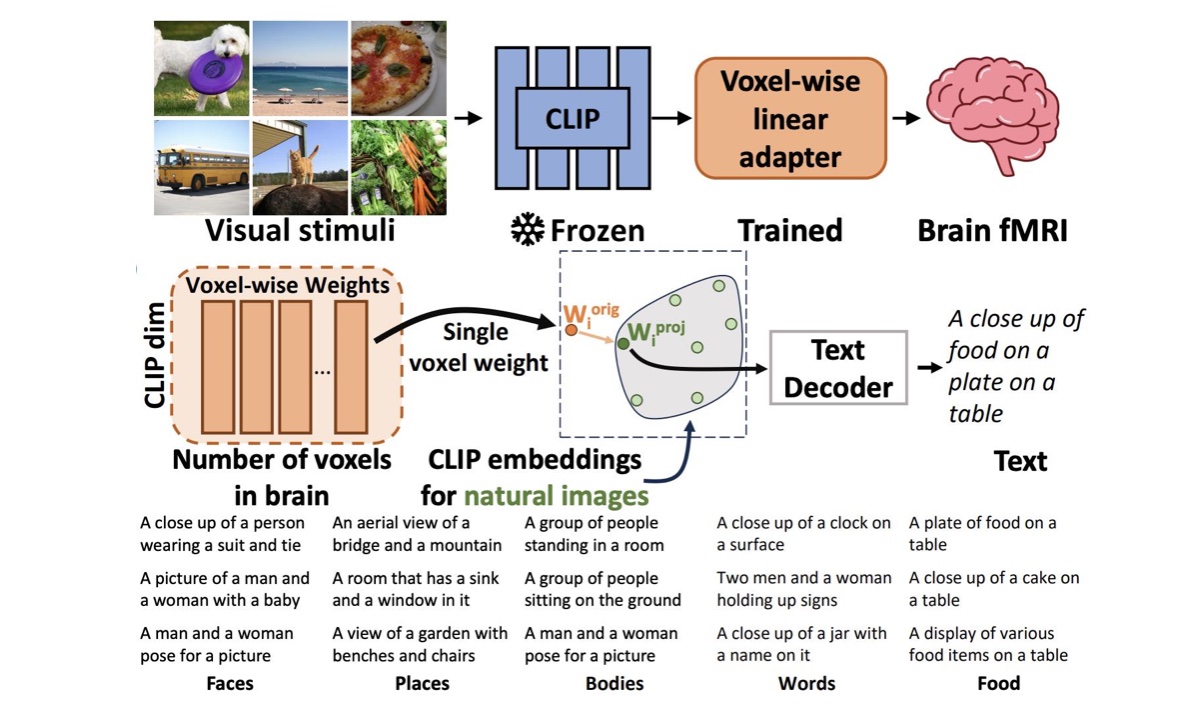
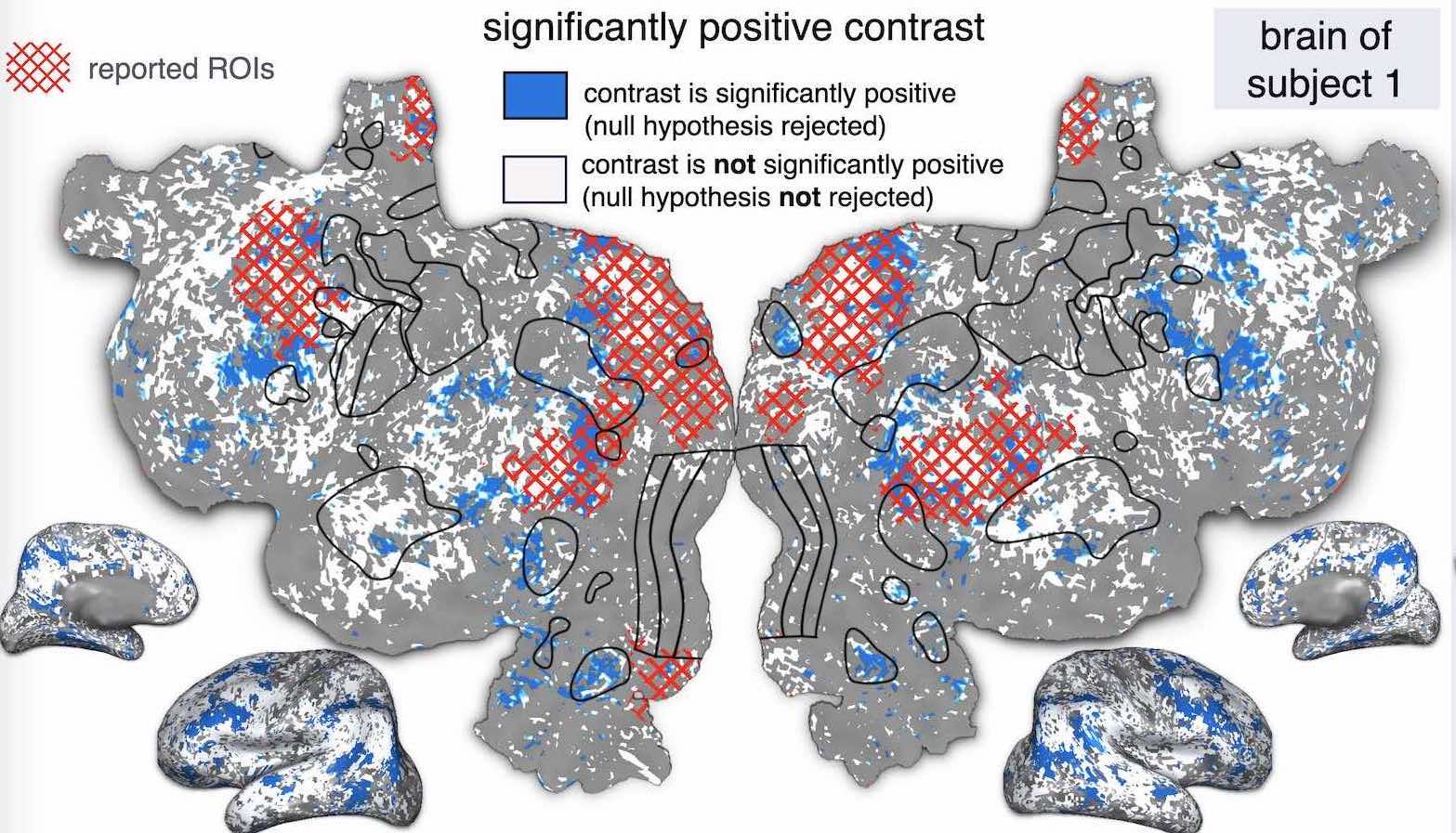
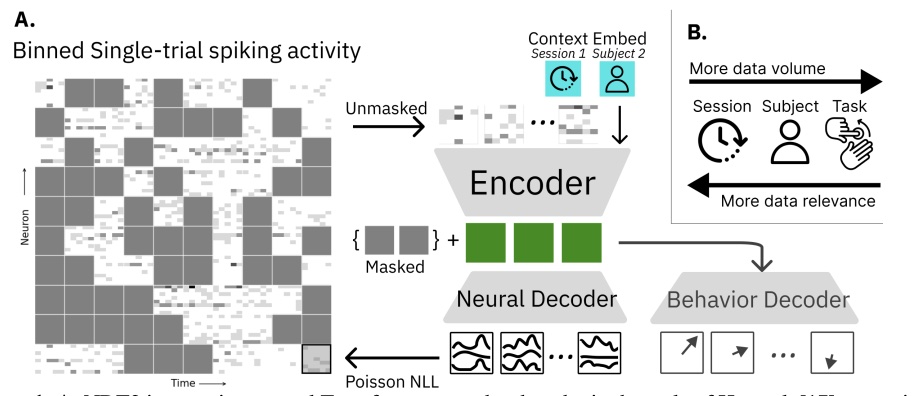
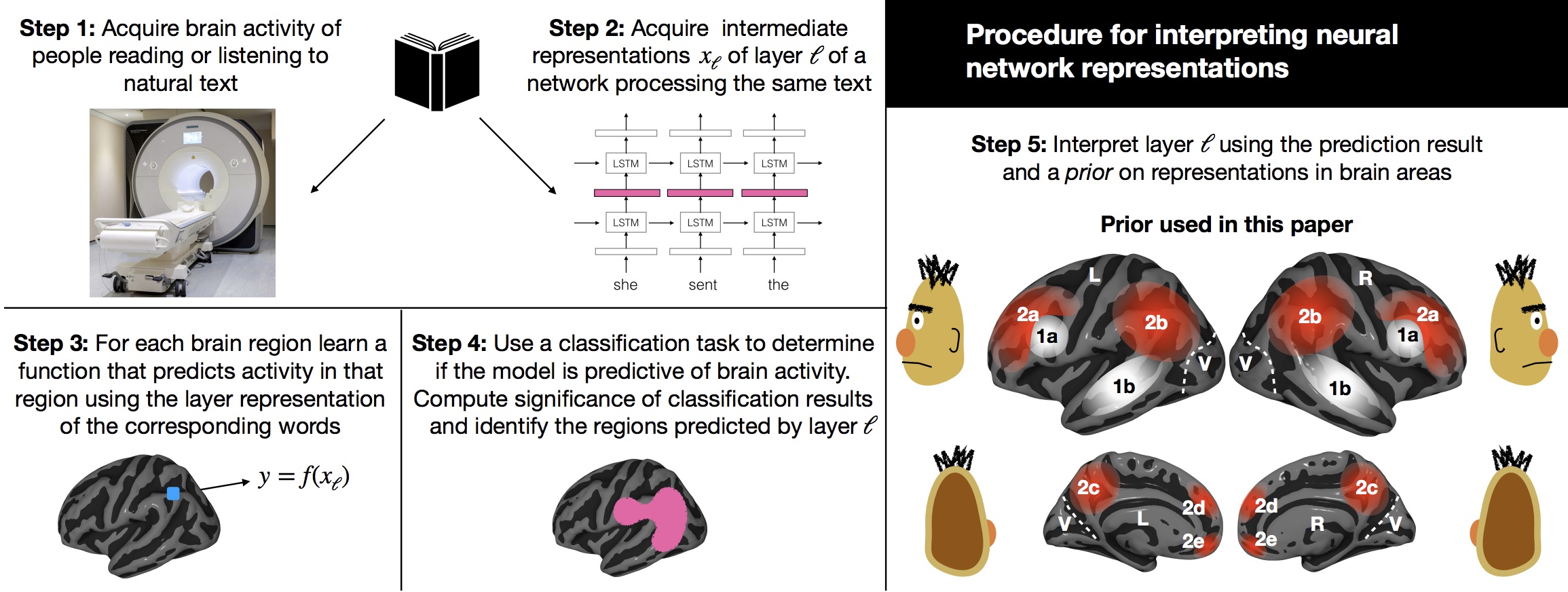
Peer reviewed publications
M. Henderson, M. Tarr, L. Wehbe.
Trends in Neurosciences, 2025.
journal
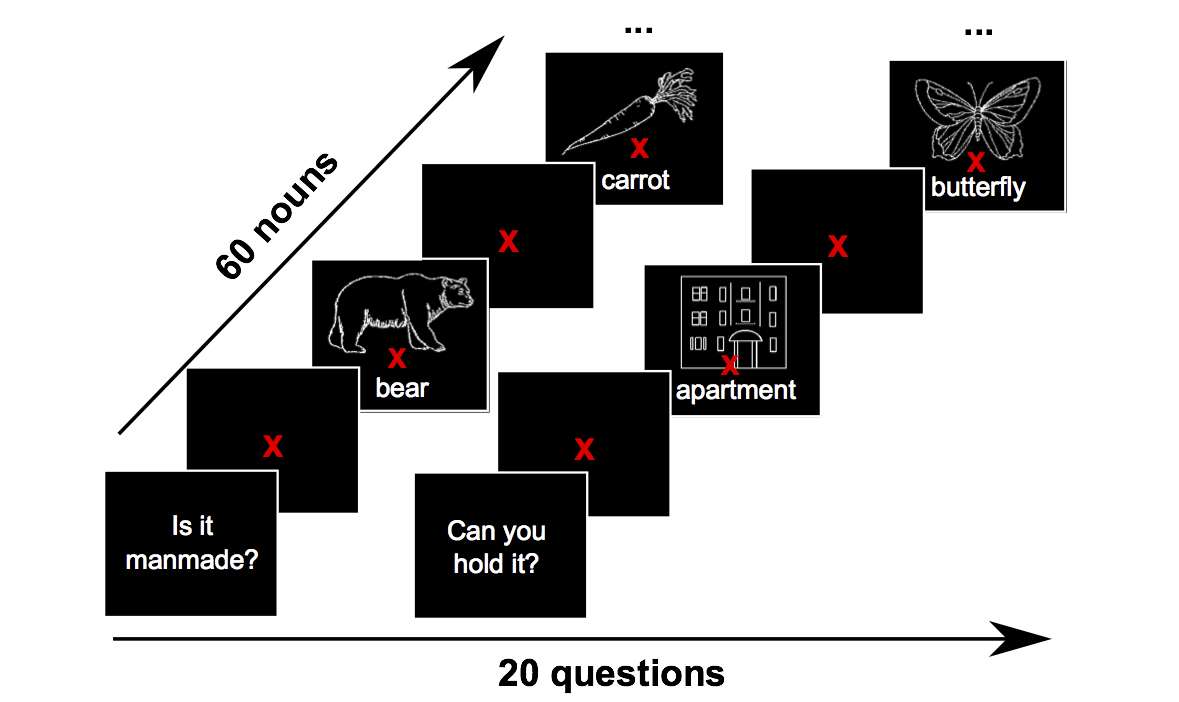
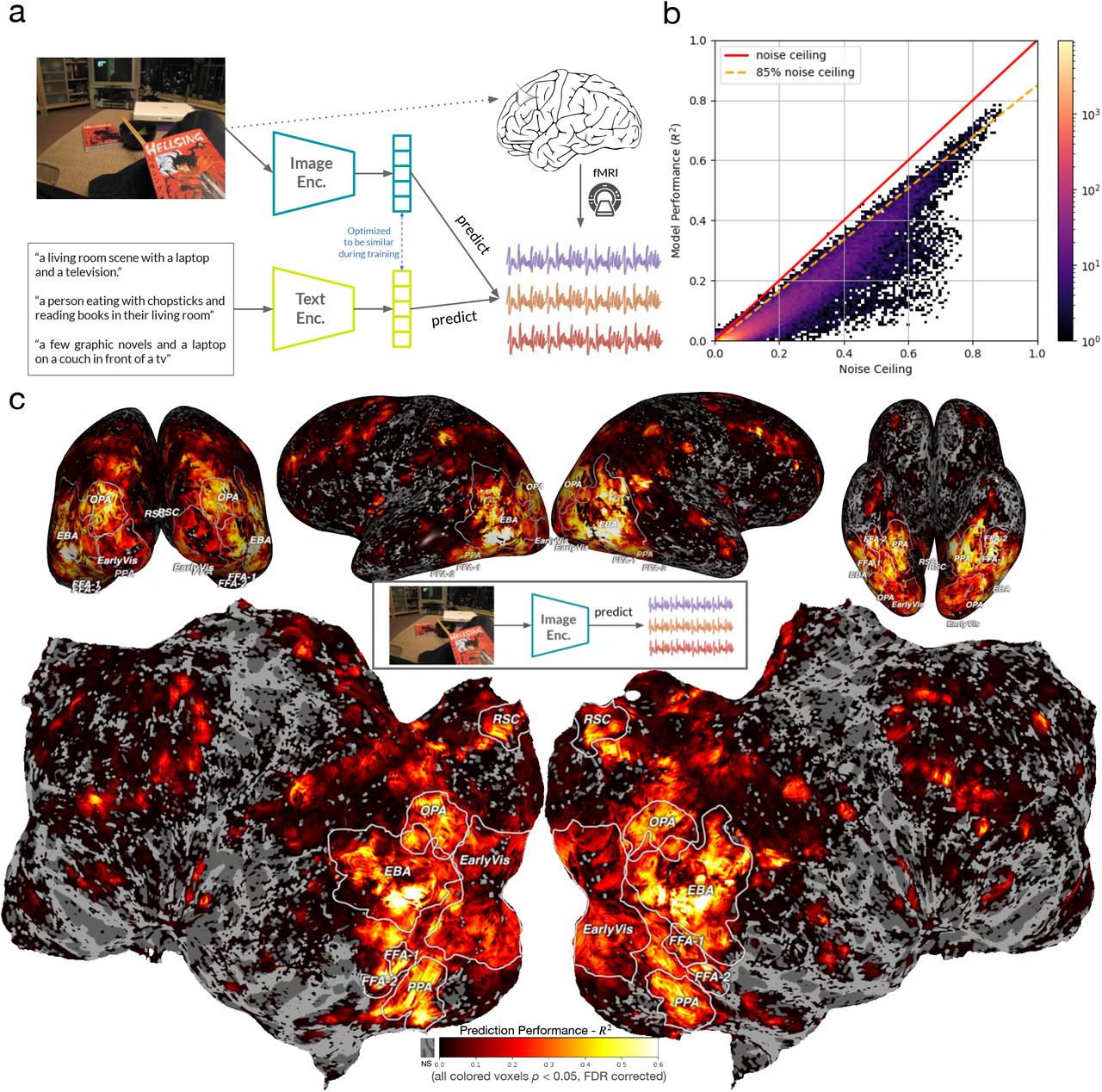
A. Luo, J. Yeung, R. Zawar, S. Dewan, M. Henderson, L. Wehbe*, M. Tarr*.
International Conference on Learning Representations (ICLR), 2025.
paper
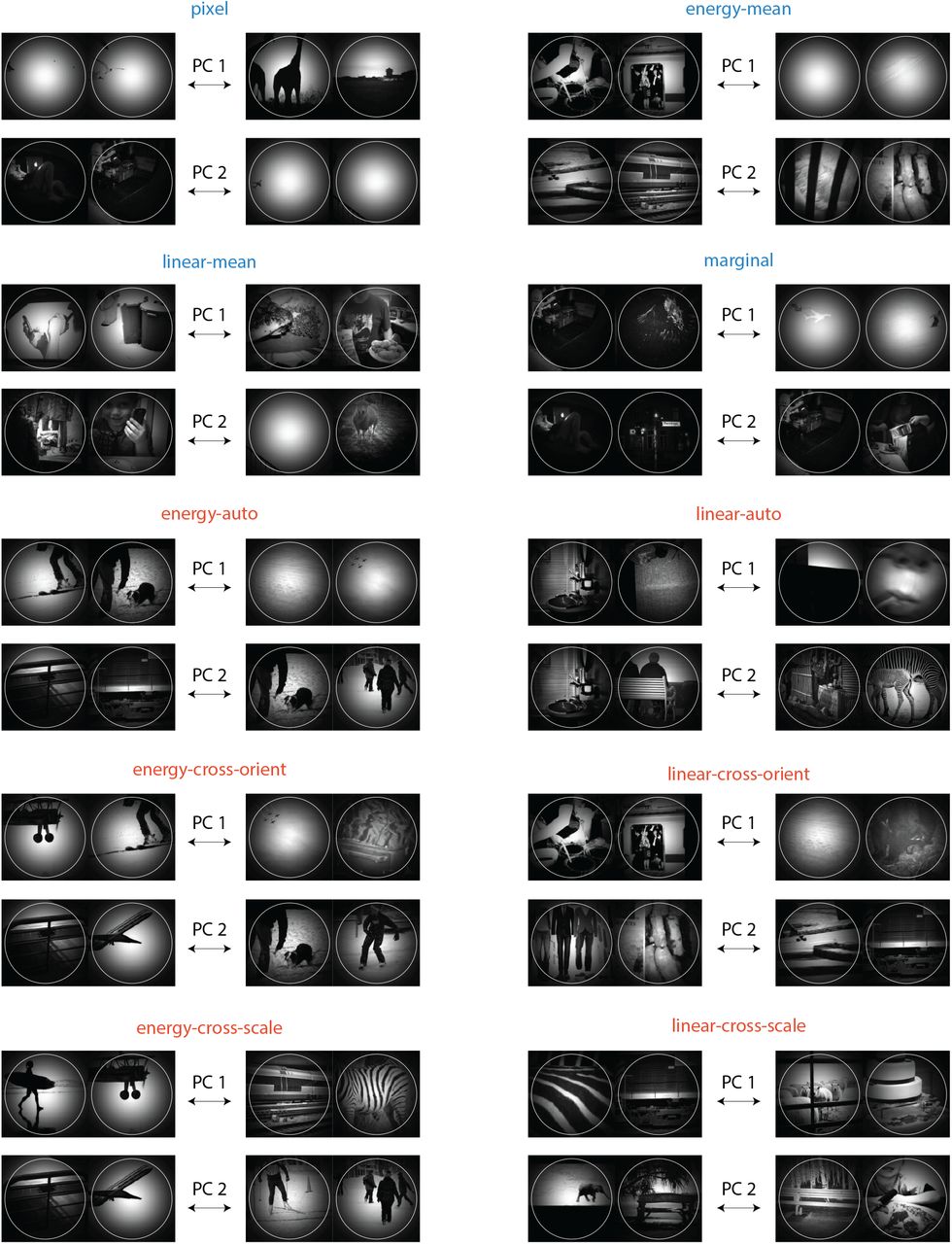
Y. Zhou, E. Liu, G. Neubig, M. Tarr, L. Wehbe.
Neural Information Processing Systems (NeurIPS), 2024.
paper presentation arXiv
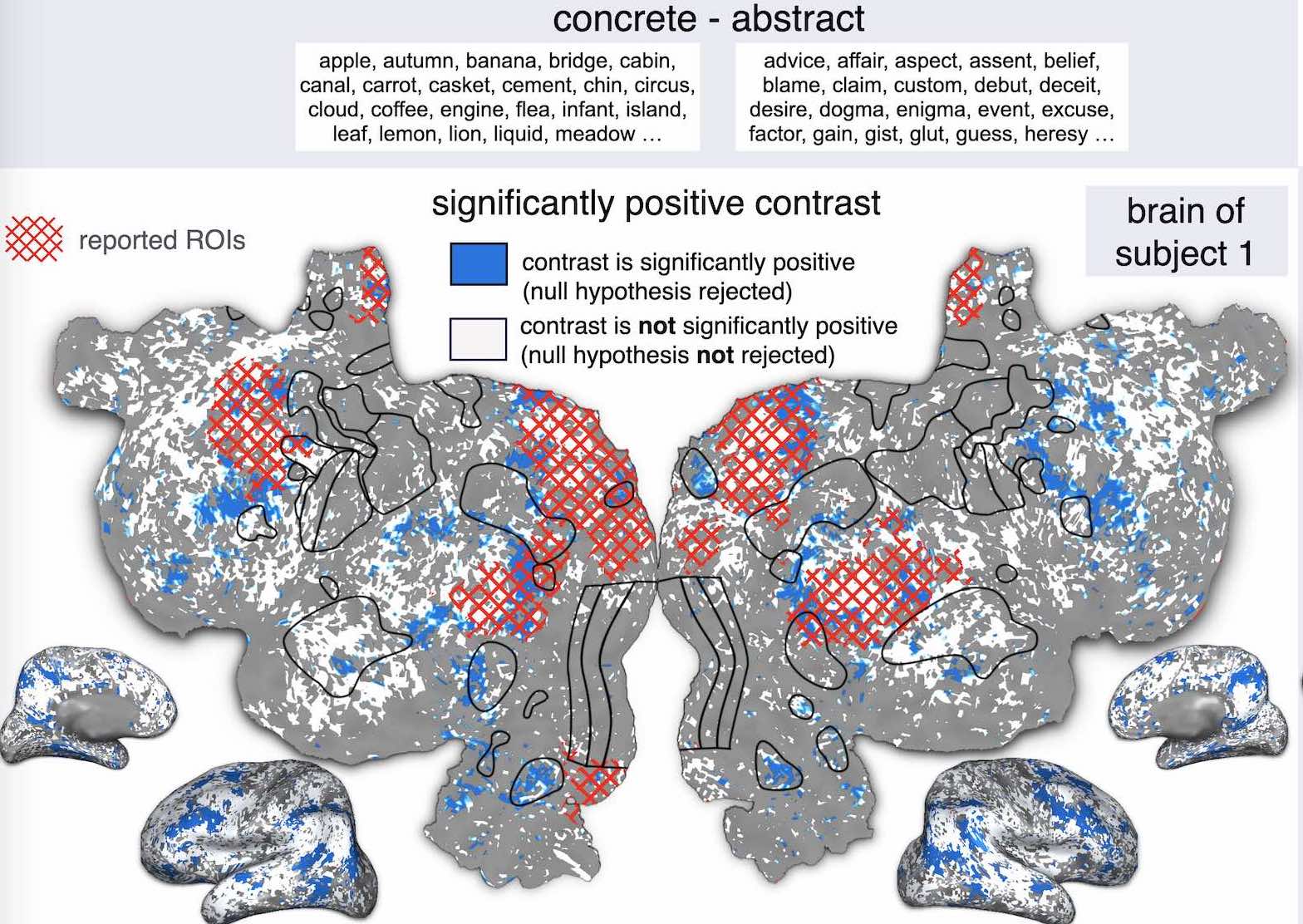
A. Luo, M. Henderson, Michael J Tarr, L. Wehbe.
International Conference on Learning Representations (ICLR), 2024.
paper
S. Jain, V. Vo, L. Wehbe, A. Huth.
Neurobiology of Language, 2024.
journal
P. Herholz, E. Fortier, M. Toneva, N. Farrugia, L. Wehbe, V. Borghesani.
Neurons, Behavior, Data analysis, and Theory, 2023.
journal
M. Khosla, L. Wehbe.
in review.
bioRxiv
M. Toneva, J. Williams, A. Bollu, C. Dann, L. Wehbe.
Proceedings of the Conference on Causal Learning and Reasoning (CLeaR) 2022.
conference arXiv
J. Williams, L. Wehbe.
in review.
arXiv
S. Ravishankar, M. Toneva, L. Wehbe.
Frontiers In Computational Neuroscience , 2021.
journal
X. Zha, L. Wehbe, R. Sclabassi, Z. Mace, Y. Liang, A. Yu, J. Leonardo, B. Cheng, T. Hillman, D. Chen, C. Riviere
IEEE Transactions on Medical Robotics and Bionics, 2020.
journal website bibtex
L. Wehbe, A. Fyshe, T. Mitchell
Human Language: from Genes and Brains to Behavior, MIT Press, 2019.
book
B. Murphy, L. Wehbe, A. Fyshe
Language, Cognition, and Computational Models, Cambridge University Press, 2018.
book
L. Wehbe, A. Ramdas, R. C. Steorts, C. R. Shalizi
Annals of Applied Statistics, 2015.
pdf journal website arXiv bibtex
brain activity during reading"
L. Wehbe, A. Vaswani, K. Knight, T. Mitchell
Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP, long paper).
supporting website pdf conference talk video bibtex
different story reading subprocesses"
L. Wehbe, B. Murphy, P. Talukdar, A. Fyshe, A. Ramdas, T. Mitchell
PLOS ONE, 2014.
supporting website (data) journal website pdf supplementary material bibtex
G. Sudre, D. Pomerleau, M. Palatucci, L. Wehbe, A. Fyshe, R. Salmelin and T. Mitchell
NeuroImage, 2012.
journal website pdf bibtex
posters, workshop papers and demos
L. Wehbe*, A. Nunez-Elizalde*, A. Huth, F. Deniz, N. Bilenko, J. Gallant.
Cognitive Computational Neuroscience, CCN 2017 poster.
L. Wehbe, A. Huth, F. Deniz, M. Kieseler, J. Gallant.
Organization for Human Brain Mapping, OHBM 2017 poster and talk..
NIPS 2016 demonstration track.
online engine
L. Wehbe, I. Blank, K. Mahowald, R. Furell, S. Piantadosi, H. Tily, J. Gallee, A. Vishnevetsky, E. Gibson, N. Kanwisher, E. Fedorenko
Society for the Neurobiology of Language, SNL 2015 poster.
L. Wehbe, A. Vaswani, K. Knight, T. Mitchell
Society for the Neurobiology of Language, SNL 2014 poster.
L. Wehbe, P. Talukdar, B. Murphy, A. Fyshe, G. Sudre, T. Mitchell
NIPS 2012 Workshop in Machine Learning and Interpretation in NeuroImaging, paper, chosen for oral presentation.
A. Fyshe L. Wehbe, B. Murphy, T. Mitchell
NIPS 2012 Workshop in Machine Learning and Interpretation in NeuroImaging, paper.
selected press coverage
- Brain scans reveal the areas that light up when we look at food, New Scientist 2022
- Google, Intel, MIT, and more: a NeurIPS conference AI research tour, ZDNET 2019
- Interviewed for the book Artificial Intelligence: Teaching Machines to Think Like People, O'Reilly Media 2017
- Harry Potter reveals secrets of the brain, Science News for Students 2015
- Lab-coated Muggles use Harry Potter to study brain, Associated Press 2014
- Reading Harry Potter Provides Clues to Brain Activity, Time 2014
- How our Brains Process Books, Scientific American 2014
- Scans map the brain as people read ‘Harry Potter’, Futurity 2014
- Reading Leaves a Dramatic Imprint on the Brain, Bioscience Technology 2014
- Episode 29, NSF Science Now
- What Harry Potter Can Teach Us About Neuroscience, Huffington Post 2014
- Reading Harry Potter: Carnegie Mellon Researchers Identify Brain Regions That Encode Words, Grammar, Character Development, CMU press release 2014