Data Visualization Tool Provides Richer Understanding of a Cell's Nucleus Nucleome Browser Gives Researchers a Map-Like Application To View Data From Multiple Sources
Aaron AupperleeThursday, September 1, 2022Print this page.
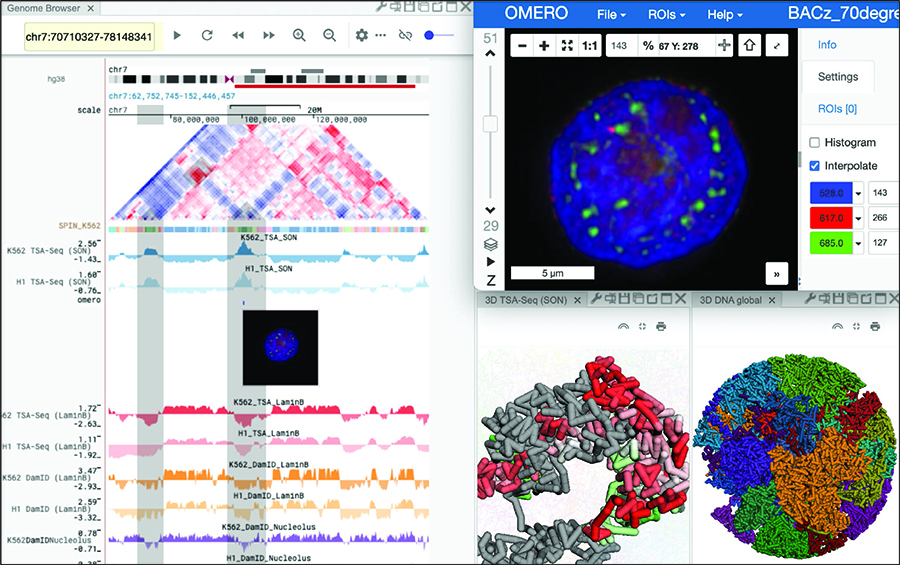
Researchers in the School of Computer Science's Computational Biology Department have developed a tool that aims to help scientists gain a fuller and deeper understanding of the structure and function of a cell's nucleus.
The Nucleome Browser is a data visualization and exploration platform allowing researchers and scientists to integrate and interact with multimodal data types, including genomic and imaging datasets. The tool will make it easier to access, use and share data, and allow for simultaneous and synchronized visualization of complex datasets related to the structure and function of the nucleus — considered the brain of the cell.
Jian Ma, the Ray and Stephanie Lane Professor of Computational Biology at Carnegie Mellon University and a lead researcher on the project, likened the tool to online mapping applications like Google Maps that integrate traffic data, business hours, restaurant reviews and other information with directions. With the open-sourced Nucleome Browser, scientists and researchers can view integrated data and annotations from many sources.
"We can see where cellular components are in the nucleus and how they interact with what's around them spatially. We can navigate imaging and genomic data together with the potential to produce new hypotheses and reveal new connections that we haven't seen before," Ma said.
This tool was developed by Xiaopeng Zhu and Yang Zhang in Ma's lab, in collaboration with researchers from the University of Illinois and the University of Dundee in Scotland. Their work, "Nucleome Browser: An Integrative and Multimodal Data Navigation Platform for 4D Nucleome," appeared in the August issue of Nature Methods.
Genomic data describes the makeup of the chromosomes inside a cell's nucleus and is typically represented as one- or two-dimensional data. Imaging data and chromosome structure models, which show how chromosomes are folded and where they are located inside the nucleus, are three-dimensional in nature. The Nucleome Browser combines these multimodal data into a unified visualization.
"Researchers can get a more unbiased and complete view of how the cell nucleus is structured," Ma said.
The browser hosts data from the National Institutes of Health's 4D Nucleome program, for which Ma's lab leads a multi-institutional center headquartered at CMU, and several other well-known and often-used data portals and applications. The new web-based platform creates an ecosystem for other users to contribute complementary and interactive applications for a wide range of biomedical contexts. The tool will be used by computational biologists, cell biologists, genomicists and even clinicians interested in exploring the link between genetic information, chromosome structure and diseases.
Aaron Aupperlee | 412-268-9068 | aaupperlee@cmu.edu
